The Components of GP2’s Second Data Release

In April 2022, GP2 announced the second data release on the Terra platform in collaboration with AMP® PD. This release contains data from both complex and monogenic GP2 networks. The complex disease data now consists of a total of 8,644 genotyped participants (5,249 PD, 3,395 non-PD). New to this release are Movement Disorders Genotypes and Phenotypes – Queen Square Brain Bank (MDGAP-QSBB), a United Kingdom brain bank, and SYNAPS Study – Kazakhstan (SYNAPS-KZ), a PD cohort from Kazakhstan. Additional CORIELL samples are also added in this release. The monogenic disease data consists of 235 whole genome sequenced (WGS) participants with PD from the PDGENEration cohort, which were selected based on set criteria for having a suspected monogenic cause of PD from the Monogenic disease network.
The key difference between genotyping microarray and WGS data is in the number of genetic markers that are detected during the genotyping process. WGS provides a comprehensive view of the genome by potentially interrogating all 3.2 billion base-pairs in the human genome, while genotyping with a targeted microarray (such as GP2’s custom NeuroBooster array) interrogates a more targeted number of up to 1.9 million region-tagging markers per sample. WGS are better suited for analyses investigating rare genetic variation, while genotypes imputed to a well-matched reference panel are a scalable and efficient solution for studying more common genetic variation.
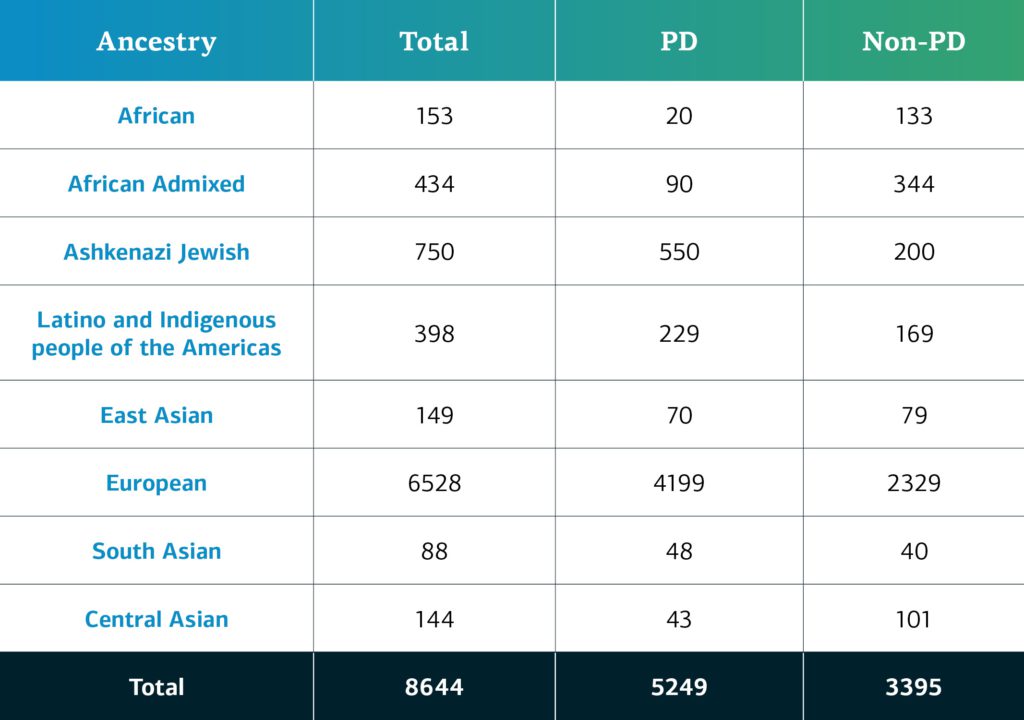
Genetically-determined ancestry of complex disease GP2 participants is broken into nine ancestry groups; the table below details the genetically-determined ancestry of complex disease participants in GP2 release 2 that have been passed quality control and been imputed.
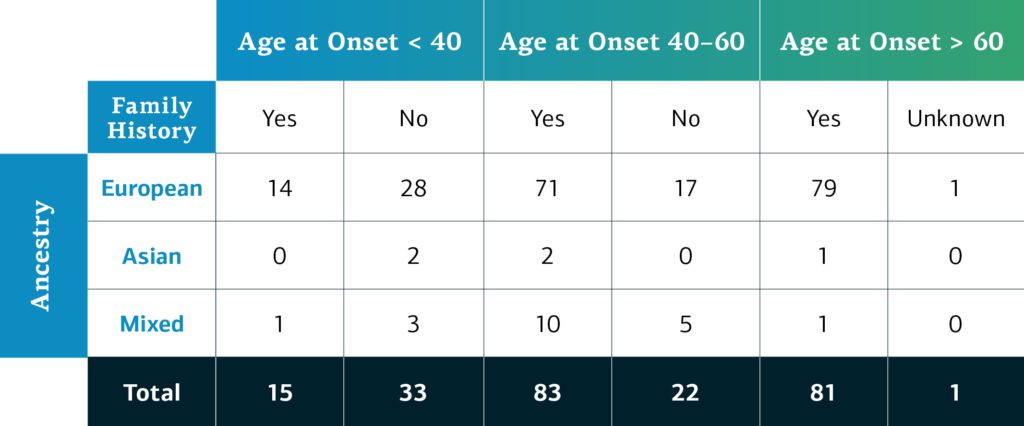
Genetically-determined ancestry of monogenic disease GP2 participants is broken into four main ancestry groups at this time, as this is the first available data from the monogenic network. As more data is processed, more diverse samples will become available. The table below details the genetically-determined ancestry of monogenic disease participants in GP2 release 2 by age at onset and family history.
Future data releases will continue to grow the diversity of participants available. You can check out our dashboard to see our progress.
In addition to including the first WGS data, we have included an additional new data type: probabilistic copy number variant (CNV) calls for all genotyped samples passing quality control (gene-level plus 250kb flanking regions). CNV refers to variation in the number of times a certain stretch of DNA is repeated. This variation may have come about through deletions, insertions, or other events and can potentially provide more information about how structural variation affects disease risk. The pipeline used to produce the probabilistic CNV calls can be found on the GP2 Github. This is currently a work in progress and will improve as we include more data and make adjustments to the pipeline. Consider this CNV data “hypothesis generating”. Usage notes will be included in the blog post covering the first stable release scheduled for next quarter.
This release contains WGS data from the monogenic disease network in addition to the NeuroBooster array genotyped complex disease data. More information on the structure of the complex disease genotype and clinical data is detailed in the blog post ‘The Components of GP2’s First Data Release’ as well as in the README which is updated at each release and is available on the official GP2 Terra workspaces. The monogenic PD WGS data is also detailed in the same README.
We are excited to make this beta release available to the PD research community and there is much more to come in the near future!
This blog was jointly authored by Hampton Leonard, Mike Nalls, Dan Vitale, Yeajin Song, Kristin Levine, Mary Makarious, Zih-Hua Fang and Peter Heutink. Please visit GP2’s Complex Disease – Data Analysis Working Group and Monogenic – Data Analysis Working Group to learn more about their background.
Check out our other data releases.