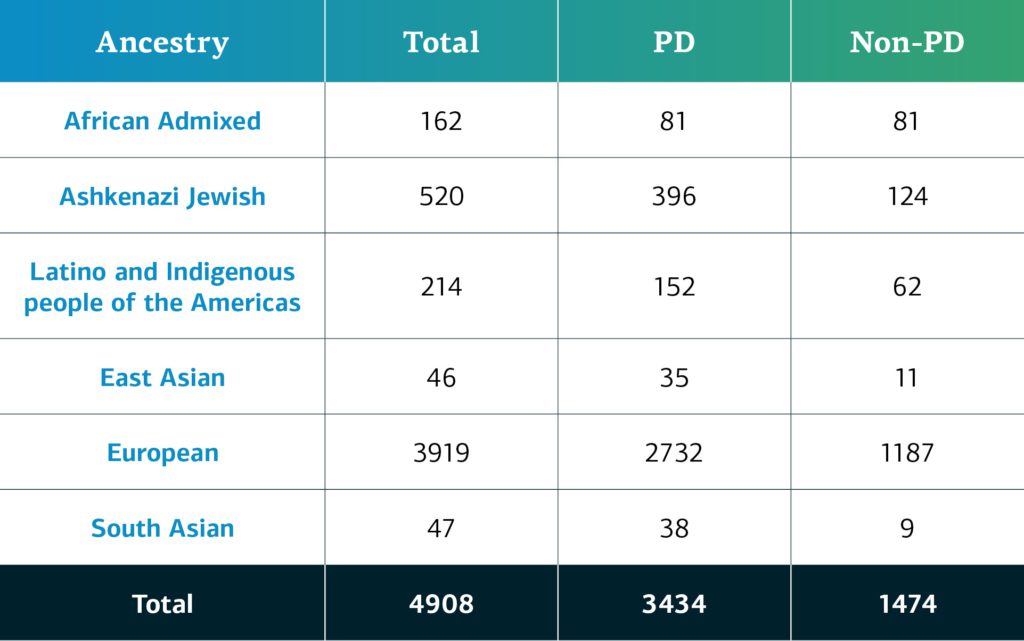
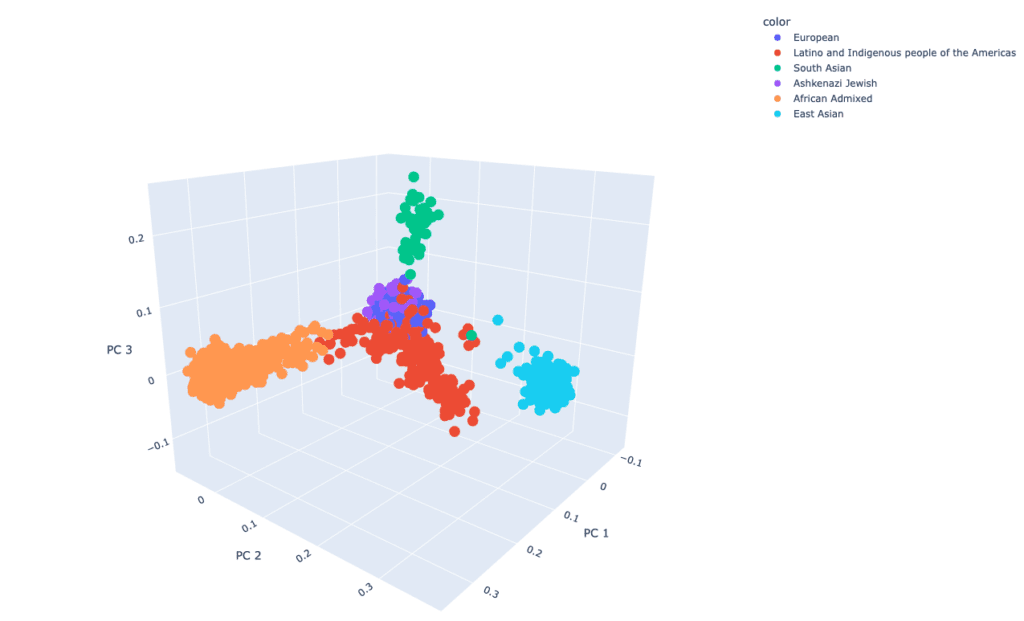
In December 2021, GP2 announced its first GP2 data release on the AMP® PD platform. The data available consists of 4,908 participants (3,434 PD, 1,474 non-PD) from the CORIELL, Baylor College of Medicine (BCM), and BCM-University of Maryland (UMD) cohorts. Genetically-determined ancestry of GP2 participants is broken into eight ancestry groups; the table below details the genetically-determined ancestry of participants in GP2 release 1.
Future data releases will continue to grow the diversity of participants available. You can check out our dashboard to see our progress.
Genotype data
All data was genotyped on the Illumina NeuroBooster Array, an array specially designed for neurodegenerative disease variants in diverse populations. Data is processed according to standard Illumina protocols and quality control of data is handled by GP2 using our open source QC pipeline. Data is then imputed using TOPMed within each separate ancestry group. All known duplicates are removed, but related samples remain in order to facilitate flexibility in community analyses.
For more information on what is included in the QC pipeline, please follow the workflow detailed on the associated GitHub page.
Clinical data
Clinical data was harmonized to form a unified GP2 clinical dataset. Minimal requirements for clinical data include demographics, recruitment category, and family history. For more information on the GP2 core data requirement levels, please refer to the “Clinical Data Core Data Set” document in the resources section on our website.
Data will be accessed through the Terra platform.
To find out how to access GP2 data, please read our blog, “Applying for GP2 Data Access on the AMP® PD Platform”. The following data products are available on Terra for GP2 release 1:
- Tier 1 data access
- Summary statistics from the most recent Parkinson’s disease GWAS (excluding 23andMe samples from Nalls et al. 2019).
- Tier 2 data access
- Raw genotypes: PLINK2 .pgen files for each ancestry group for all samples passing quality control prior to imputation
- Imputed genotypes: PLINK2 .pgen files imputed using the TOPMed reference panel (passing minimum QC filtering detailed in the readme file accompanying the data release)
- Meta data: contains information relating to QC outputs and ancestry prediction
- Clinical data: clinical data fields and relevant clinical data dictionary
We hope this becomes a valuable resource for the Parkinson’s disease research community. We are continuously improving our processes and have many more exciting things to come.
This blog was jointly authored by Hampton Leonard, Mike Nalls, Yeajin Song, and Dan Vitale. Please visit GP2’s Complex Disease – Data Analysis Working Group page to learn more about their background.
Check out our other data releases.