In October 2022, GP2 announced the third data release on the Terra platform in collaboration with AMP® PD. This release is packed full of new data and resources and includes almost twice as many samples as the previous release.
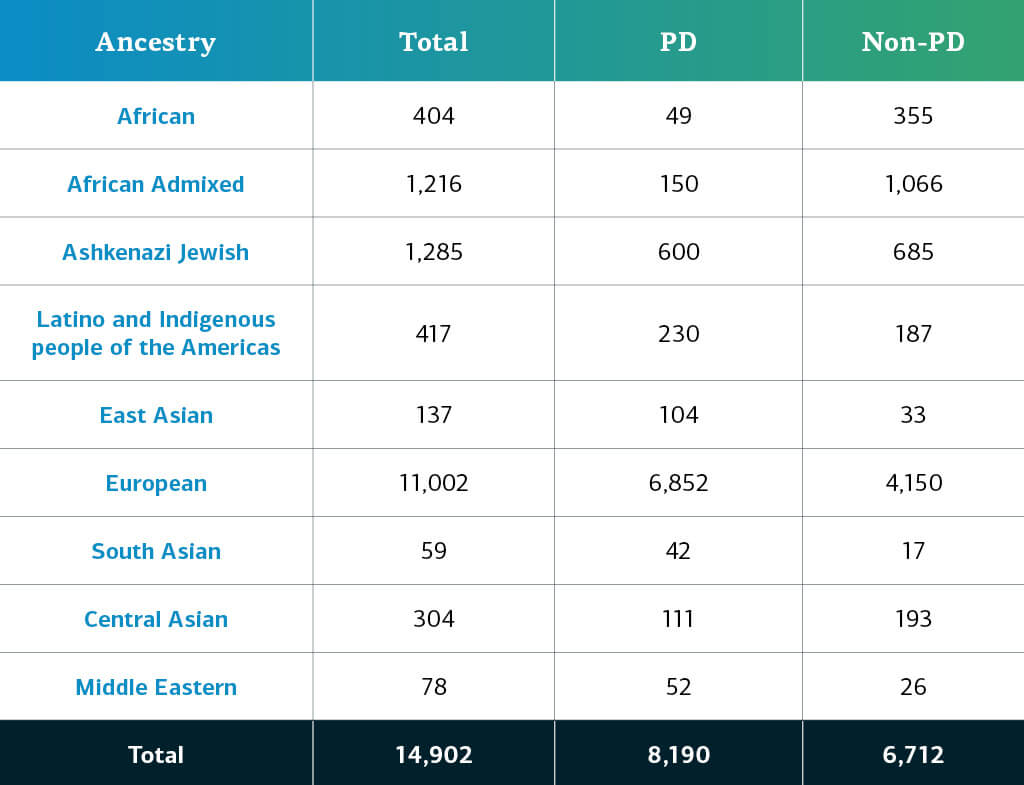
This release includes 6,330 additional new complex disease participants, adding to the previous releases from the Complex and Monogenic Networks. The complex disease data now consists of a total of 14,902 genotyped participants (8,190 PD; 6,712 non-PD). New cohorts to this release are:
- The Australian Parkinson’s Genetics Study (APGS)
- The Fox Investigation for New Discovery of Biomarkers (BioFIND), an observational clinical study of PD
- Participating sites from The Black and African American Connections to Parkinson’s Disease (BLAACPD-KPM, BLAACPD-RUSH, BLAACPD-UAB, BLAACPD-UC), a PD cohort of Black and African American individuals
- Koc University GP2 Cohort (KOC), a Turkey-based cohort
- The LRRK2 Cohort Consortium (LCC), a PD cohort of LRRK2 mutation carriers
- Movement Disorders Genotypes and Phenotypes – King’s College London (MDGAP-KINGS), a UK-based brain bank
- New Zealand Parkinson’s Programme (NZP3)
- Parkinson’s Genes and Environment Study (PAGE)
- The Parkinson’s Progression Markers Initiative (PPMI)
- Systemic Synuclein Sampling Study (S4), a PD biomarker study of alpha-synuclein
Genetically-determined ancestry of complex disease GP2 participants is broken into ten ancestry groups (the nine groups below plus a small number of Finnish Europeans); the table below details the genetically-determined ancestry of complex disease participants in this release have passed quality control and been imputed. These numbers include samples from previous releases that have been reclustered using the new cluster file and gone through quality control along with the newly genotyped and shared samples unique to this current release.
Future data releases will continue to grow the diversity of participants available. You can check out our dashboard to see our progress.
The primary deliverable of this dataset is including more samples with array-based and imputed genotype data plus additional clinical and genomic metadata. This data has been clustered using a custom GP2 genotype clustering file (available in the utils directories under both tier 1 and tier 2 data access). This cluster file is based on 2,793 samples across 6 ancestry groups representative of the diverse genetic ancestries comprising this GP2 data release as well as an enrichment of 420 Gaucher disease cases to better capture variants of interest in the GBA risk gene. The cluster file is also available for download via the GP2 GitHub repository if you would like to apply this to data outside of GP2.
We also have updated the copy number variant (CNV) calls for all genotyped samples passing quality control (gene-level plus 250kb flanking regions). CNV refers to variation in the number of times a certain stretch of DNA is repeated. This variation may have come about through deletions, insertions, or other events and can potentially provide more information about how structural variation affects disease risk. The pipeline used to produce the probabilistic CNV calls can be found on the GP2 Github. These results are stratified by genetic ancestries in the tier 2 access workspace. These probabilistic CNVs are a great starting point to prioritize samples carrying potential insertions, deletions or duplications in genes of interest for follow-up studies and further analyses.
More information on the structure of the complex disease genotype and clinical data is detailed in the blog post ‘The Components of GP2’s First Data Release’ as well as in the README that has been updated for this release and is available on the official GP2 Terra workspaces. The monogenic PD WGS data is also detailed in the same README. More monogenic WGS data will become available next release.
Preview of next release includes (targeting holiday season, calendar Q1-2023):
- More genotype data
- More WGS data
- More clinical metadata
- Updated analysis code and tools
- Additional summary statistics for risk and age at onset GWAS studies
This blog was authored by members of the Complex Disease – Data Analysis Working Group: Hampton Leonard, Mike Nalls, Dan Vitale and Mary Makarious; Matthew Korestsky from the National Institutes of Health; Kristin Levine from Data Tecnica International/National Institutes of Health; and Zih-Hua Fang and Peter Heutink, members of the Monogenic – Data Analysis Working Group.
Check out our other data releases.





