In May 2023, GP2 announced the fifth data release on the Terra platform in collaboration with AMP® PD.
This release includes 7,462 additional new complex disease participants and 487 new monogenic disease participants, adding to the previous releases from the Complex and Monogenic Networks.
- The complex disease data (genotypes) now consists of a total of 24,935 genotyped participants (12,728 PD cases, 10,533 Controls, and 1,674 ‘Other’ phenotypes).
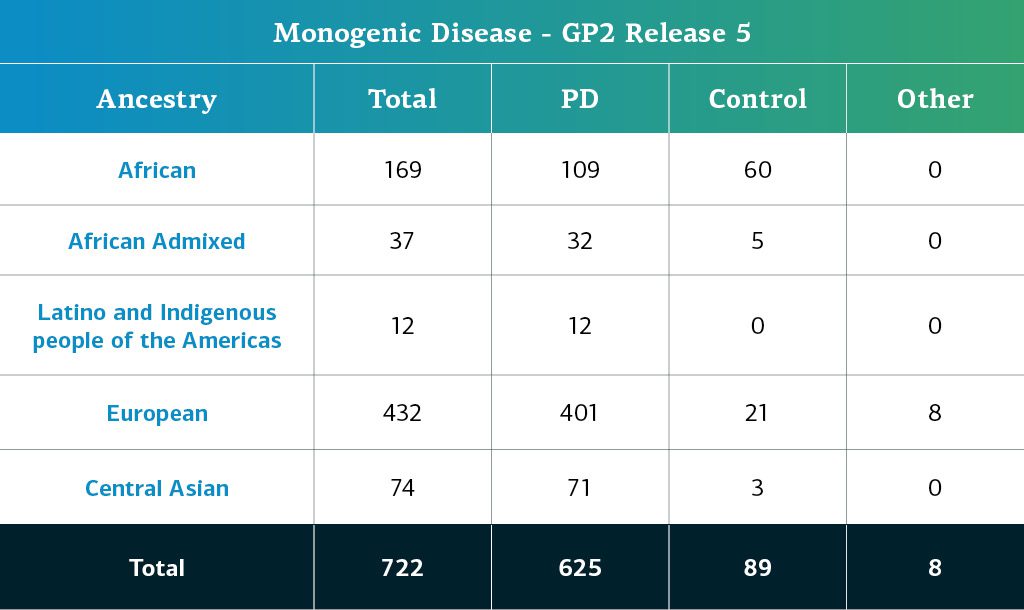
- The monogenic disease data (whole genome sequences) now consists of a total of 722 sequenced participants.
Individual-Level Data:
New cohorts added to GP2 in this release are:
- Arizona Brain Bank Brain and body donation program (BBDP), a US-based brain bank
- Innate and Adaptive Immunity in Parkinson Disease (IMMUNEPD), a US-based study from the University of Alabama at Birmingham
- IPDGC Africa (IPDGCAF-NG), a Nigerian cohort
- PDGENEration (PDGNRTN), a cohort from the Parkinson’s Foundation
- Genetic study of Parkinson’s disease and related movement disorders (STELLENBOS), a South African cohort from Stellenbosch University
- Malaysian Parkinson’s Genetics Cohort (UMKLM), a Malaysian cohort from the University of Malaya
- Genotype-Phenotype Correlations in Parkinson’s Disease and Related Movement Disorders (KUL), a monogenic cohort from the University of Malaysia
- Parkinson Disease and Movement Disorders Center Biorepository (PDMDC), a monogenic cohort from Northwestern University
Genetically-determined ancestry of complex disease GP2 participants is broken into ten ancestry groups; the table below details the genetically-determined ancestry of complex disease participants in this release that have passed quality control and been imputed. These numbers include samples from previous releases that have been reclustered using the new cluster file and gone through quality control along with the newly genotyped and shared samples unique to this current release.
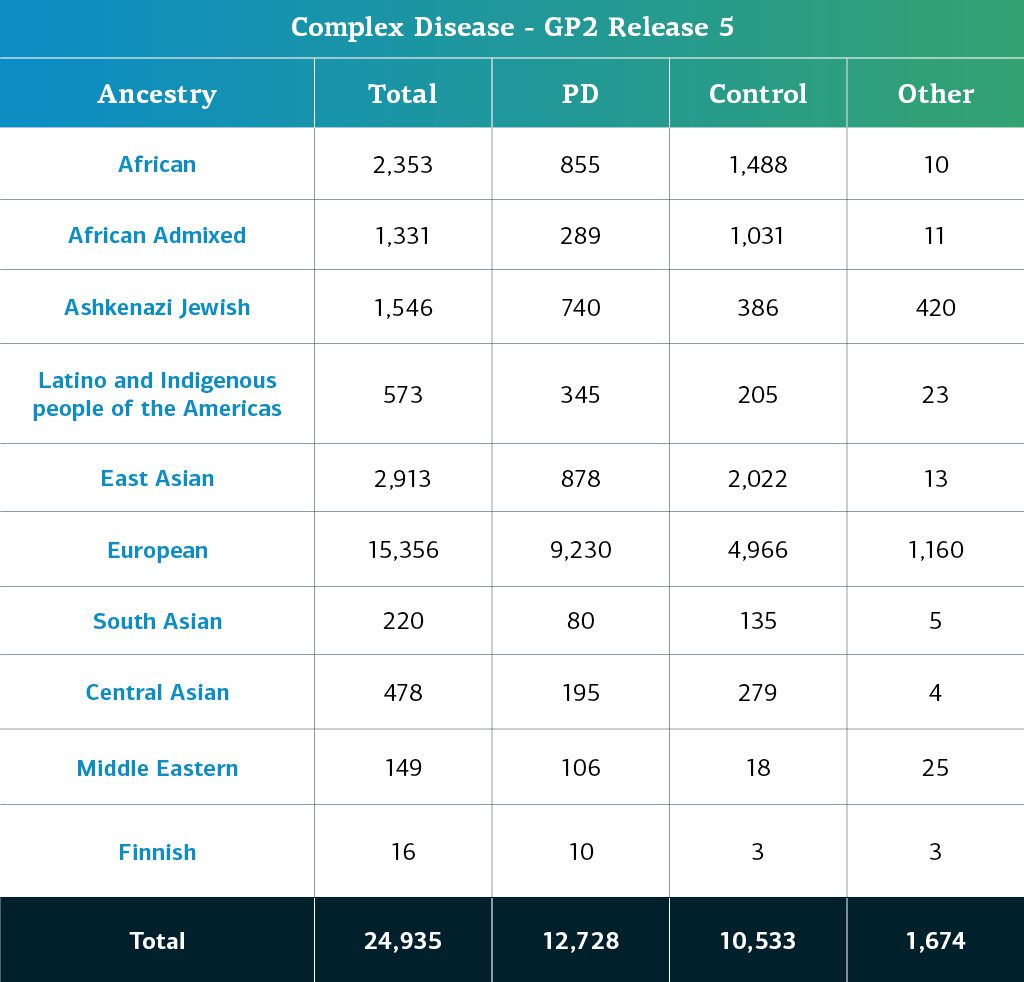
Important to note in this release: STELLENBOS is a South African cohort and displays genetic ancestry admixture that reflects that region and its history. Some of our ancestry predictions for members of this study may change as we adjust for this new population in future releases. In this release we have provided admixture estimates for all participants so that researchers can make choices about if and how to include high admixture in their analysis as part of the tier 2 data. We have also included a list of samples that we recommend removing from analysis at this stage as their high levels of admixture lead to lower confidence in their ancestry predictions from our current model.
Genetically-determined ancestry of monogenic disease GP2 participants is estimated from the same pipeline as the complex participants. The table below details the genetically-determined ancestry of monogenic disease participants in GP2 release 5.
Future data releases will continue to grow the diversity of participants available. You can check out our dashboard to see our progress.
An important update for this release is a minor change to how we determine which participants are related. We are now using KING software. This change may have updated estimates of relatedness for some of our previously released participants. With this change, a file that details the estimated relatedness thresholds will now be made available at the release for researchers to use for their own custom filtering purposes. In addition to the projected principal components, we now include principal components calculated per ancestry available for downstream analyses.
Summary Statistics:
We are pleased to announce that new summary statistics are also available through tier 1 access for this release. Kim et al 2023 are also available through our ongoing collaboration with Neurodegenerative Disease Knowledge Portal.
- Nalls et al 2019 without 23andme in hg38 (largest European PD GWAS)
- Loesch et al 2021 (largest Latin American/Amerindian PD GWAS)
- Kim et al 2023 without 23andme (Multi-ancestry meta-analysis for PD risk including European, East Asian, Latin American/Amerindians, and African Americans)
- Rizig et al 2023 without 23andme in hg38 (largest African and African admixed PD GWAS; using GP2 release 5 data)
Copy Number Variants:
Copy number variant (CNV) calls for all genotyped samples passing quality control (gene-level plus 250kb flanking regions) have been updated to include all samples in release 5. This data has been clustered using a custom GP2 genotype clustering file (available in the utils directories under both tier 1 and tier 2 data access). Both the cluster file and the pipeline used to predict the probabilistic CNV calls can be found on the GP2 Github for use with data outside of GP2. For more information regarding clustering using the custom GP2 genotyping clustering file and the copy number variant probabilistic calls, please see “The Components of GP2’s Third Data Release” on the GP2 blog.
More information on the structure of the complex disease genotype and clinical data is detailed in the blog post “The Components of GP2’s First Data Release” as well as in the README that has been updated for this release and is available on the official GP2 Terra workspaces. The monogenic PD WGS data is also detailed in the same README.
As always, please refer to the README that accompanies each GP2 release for further details regarding pipelines, data, and analyses!
On behalf of the GP2 Complex Disease Data Analysis, Monogenic Data Analysis, and Data and Code Dissemination Working Groups.