The Components of GP2’s Fourth Data Release
In February 2023, GP2 announced the fourth data release on the Terra platform in collaboration with AMP® PD. This release is packed full of new data and resources.
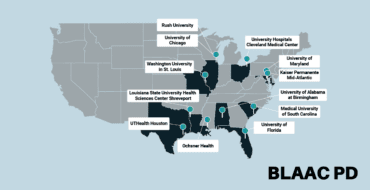
This release includes 2,583 additional new complex disease participants, adding to the previous releases from the Complex and Monogenic Networks. The complex disease data now consists of a total of 17,485 genotyped participants (9,429 PD; 6,648 Controls, and 1,408 ‘Other’). The cohorts added to GP2 in this release are:
- Canadian Open Parkinson Network (C-OPN), a collaborative network of nine Canadian universities and research centers
- Movement Disorders Genotypes and Phenotypes – Edinburgh Brain Bank (MDGAP-EBB), a UK-based brain bank
- Sydney Brain Bank (SBB), an Australian-based brain bank
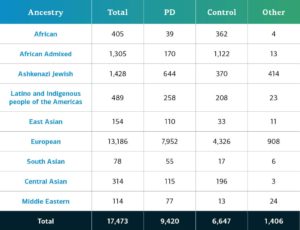
Genetically-determined ancestry of complex disease GP2 participants is broken into ten ancestry groups (the nine groups below plus a small number of Finnish Europeans); the table below details the genetically-determined ancestry of complex disease participants in this release that have passed quality control and been imputed. These numbers include samples from previous releases that have been reclustered using the new cluster file and gone through quality control along with the newly genotyped and shared samples unique to this current release.
Future data releases will continue to grow the diversity of participants available. You can check out our dashboard to see our progress.
An important update for this release is a minor change to one of the thresholds used in our QC pipeline. In order to share more samples, we have removed genotyped variants that are consistently underperforming in quality before going through the QC process. A list of the poor performing variants can be found in the meta data directory of the Tier 2 buckets and on Github. We have also used a less stringent sample call rate, changing from 0.98 to 0.95. In doing so, more samples can be used for your analyses, without sacrificing quality.
Another update for this release is an update to the clinical data. The phenotype column (“Phenotype”) has been updated to only include labels ‘PD’, ‘Control’, or ‘Other’. An additional column has been added, ‘other_pheno’, which provides a more detailed diagnosis label for participants with an ‘Other’ label. Some of these more detailed diagnosis labels include unaffected and affected individuals via targeted PD genetic recruitment, SWEDDs, and prodromal PD as well as other disorders such as essential tremor (ET), progressive supranuclear palsy (PSP), dementia with Lewy bodies (DLB), and multiple system atrophy (MSA).
Copy number variant (CNV) calls for all genotyped samples passing quality control (gene-level plus 250kb flanking regions) have been updated to include all samples in release 4. This data has been clustered using a custom GP2 genotype clustering file (available in the utils directories under both tier 1 and tier 2 data access). Both the cluster file and the pipeline used to predict the probabilistic CNV calls can be found on the GP2 Github for use with data outside of GP2. For more information regarding clustering using the custom GP2 genotyping clustering file and the copy number variant probabilistic calls, please see the “The Components of GP2’s Third Data Release” on the GP2 blog.
More information on the structure of the complex disease genotype and clinical data is detailed in the blog post ‘The Components of GP2’s First Data Release’ as well as in the README that has been updated for this release and is available on the official GP2 Terra workspaces. The monogenic PD WGS data is also detailed in the same README.