Overview
In September 2024, GP2 announced the eighth data release on the Terra and the Verily® Workbench platforms in collaboration with AMP® PD. This release includes 5,481 additional whole genome sequences and 10,454 clinical exome sequences. Additional genotyping will be provided in the following release.
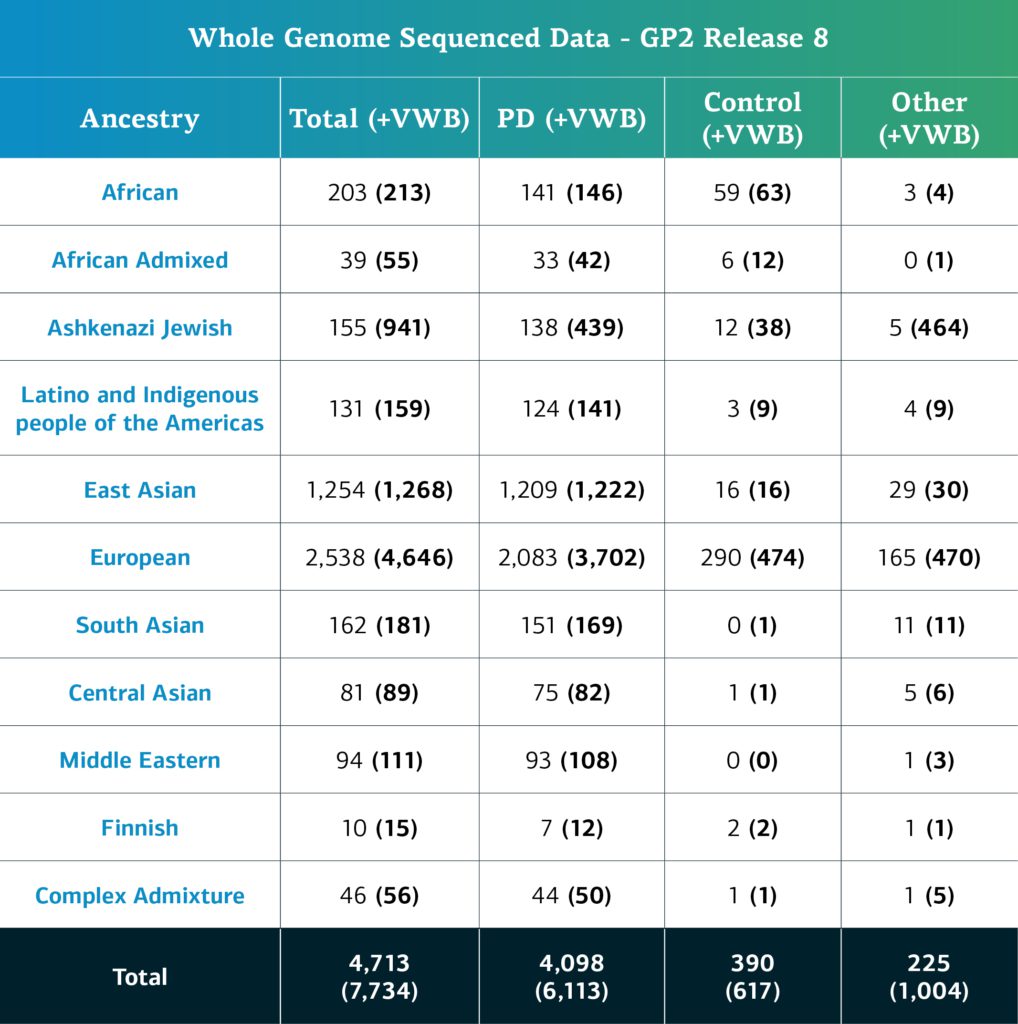
- The whole genome sequencing (WGS) data now consists of a total of 7,734 sequenced participants (6,113 PD cases, 617 Controls, and 1,004 ‘Other’ phenotypes).
- When removing the locally-restricted samples, these now consist of 4,713 participants (4,098 PD cases, 390 Controls, and 225 ‘Other’ phenotypes).
- Of note, cases recruited via the Monogenic Network are coded as ‘Other’
- Additionally, included in this WGS release is a partial release of whole genome sequences from two AMP® PD cohorts (BioFind and PPMI) that have been joint-called with GP2 WGS. Released samples can be linked back to the original AMP® PD IDs through an ID crosswalk file included with the release.
- This release also includes 10,454 joint-called clinical exome sequencing participants from the Parkinson’s Foundation.
- This release includes a total of 62,087 individuals who have core clinical data available. Among these, 16,800 individuals have deep clinical phenotyping and genetic data available.
What’s New In This Release?
- Additional GP2 whole genome sequencing samples and the joint-called variant sets including the samples from two AMP® PD cohorts (BioFind and PPMI)
- Clinical exome data from Parkinson’s Foundation
- Additional clinical data for individuals, bringing our total to 62,087 individuals who have core clinical data available
Locality-restricted GDPR samples via the Verily Viewpoint Workbench
We are continuing to pilot granting access to locally-restricted samples, otherwise known as samples governed by the General Data Protection Regulation (GDPR) policy, through our collaboration with the Verily Viewpoint Workbench.
At this time, as GP2 continues to roll out data sharing solutions for GDPR protected data, release 8 data with regional restrictions will be available to only GP2 consortium members and partners. As testing and implementation continues in 2024, this solution will be available to the broader research community. All release 8 samples can be found on Workbench. Meanwhile, all release 8 samples not governed by GDPR requirements can be found on the community workbench on Terra (like all previous releases). To gain access to the full release on VWB you must:
- Have approved GP2 Tier 2 access
- Fill out the GDPR-governed sample request form
- Be a GP2 consortium member (contributing cohort, GP2 partner, or project analyses team member)
Clinical Data
This release contains clinical data for a total of 62,087 individuals who have genetic and core clinical data available. There is deep clinical phenotyping data and genetic data for 16,800 individuals in this release. This information consists of:
- Age at diagnosis and onset
- Primary, current, and latest diagnoses
- Cognitive exams such as the Mini-Mental State Examination and the Montreal Cognitive Assessment
- Movement Disorder Society-Sponsored Revision of the Unified Parkinson’s Disease Rating Scale
- Detailed “other” phenotypes, such as Lewy body Dementia
- Cases recruited via the Monogenic network are coded as ‘Other’
Clinical Exome Sequences
Clinical exome sequencing provided by the Parkinson’s Foundation is available for 10,454 Parkinson’s Foundation samples in this release, offering analysis of the coding regions and splice junctions of 4,717 genes. This targeted sequencing aims to identify and report variants of potential clinical significance, focusing on those that align with the patient’s clinical information and family history. For more detailed information, visit the Fulgent Genetics Clinical Exome page.
Whole Genome Sequences called by DeepVariant-GLnexus
We use Google’s DeepVariant pipeline coupled with GLnexus for cohort-level variant calling. DeepVariant is a deep learning-based variant caller that outperforms existing state-of-the-art tools by accurately calling individual-level genetic variants. It also simplifies the process, enhancing accuracy and reliability.
Genetically-determined ancestry of array genotyped GP2 participants is broken into 11 ancestry groups; the table below details the genetically-determined ancestry of genotyped participants in this release that have passed quality control and been imputed. These numbers include samples from previous releases that have been reclustered using the new cluster file and gone through quality control along with the newly genotyped and shared samples unique to this current release.
Future data releases will continue to grow the diversity of participants available. You can check out our Cohort Dashboard to see our progress. For users with tier 2 access already, you can explore the data further on our Cohort Browser, expanded on in a previous blog post.
As always, please refer to the README that accompanies each GP2 release for further details regarding recommendations for quality control, pipelines, data, and analyses!