GP2’s purpose is to dramatically expand our understanding of the genetic basis of Parkinson’s and to make that knowledge globally relevant. Our path to getting there is through building a global research community with the tools and infrastructure to address emerging research needs and questions. Here, we announce the launch of our newest application – the GP2 Cohort Browser. This revolutionary tool provides an immersive, interactive platform that empowers GP2 cohort contributors and researchers to dive into the rich genomic data in an efficient and user-friendly manner. The GP2 Cohort Browser was developed to ensure a seamless and comprehensive data exploration experience for those working with our individual level GP2 genetics data. To get access to these datasets and use our tool, one must have an approved tier 2 GP2 data agreement in place, which you can begin applying for here: https://amp-pd.org/register-for-amp-pd. You can also learn more about how to apply here.
Below is an overview of the new tool that once you have signed our data use agreement, you can start exploring right away!
The browser can be found here: https://gp2-cohort-browser-dot-gp2-release-terra.uc.r.appspot.com/

Landing Page
The landing page is your first step to seeing all the web application has to offer. It sets the stage for the information you can explore and offers a sneak peek into what you can expect, from quality control measures to ancestry breakdowns.

Comprehensive Meta-data Breakdown
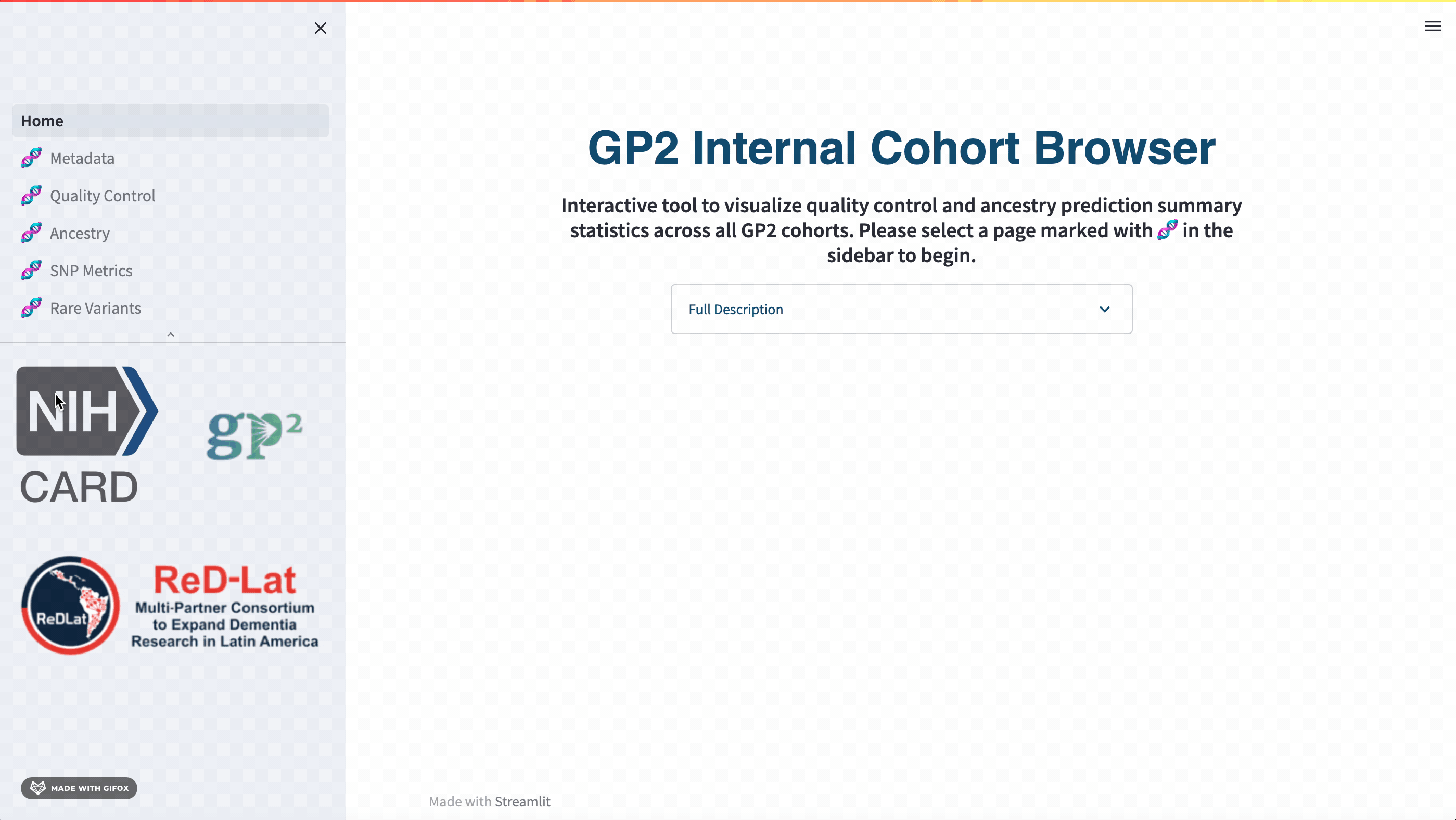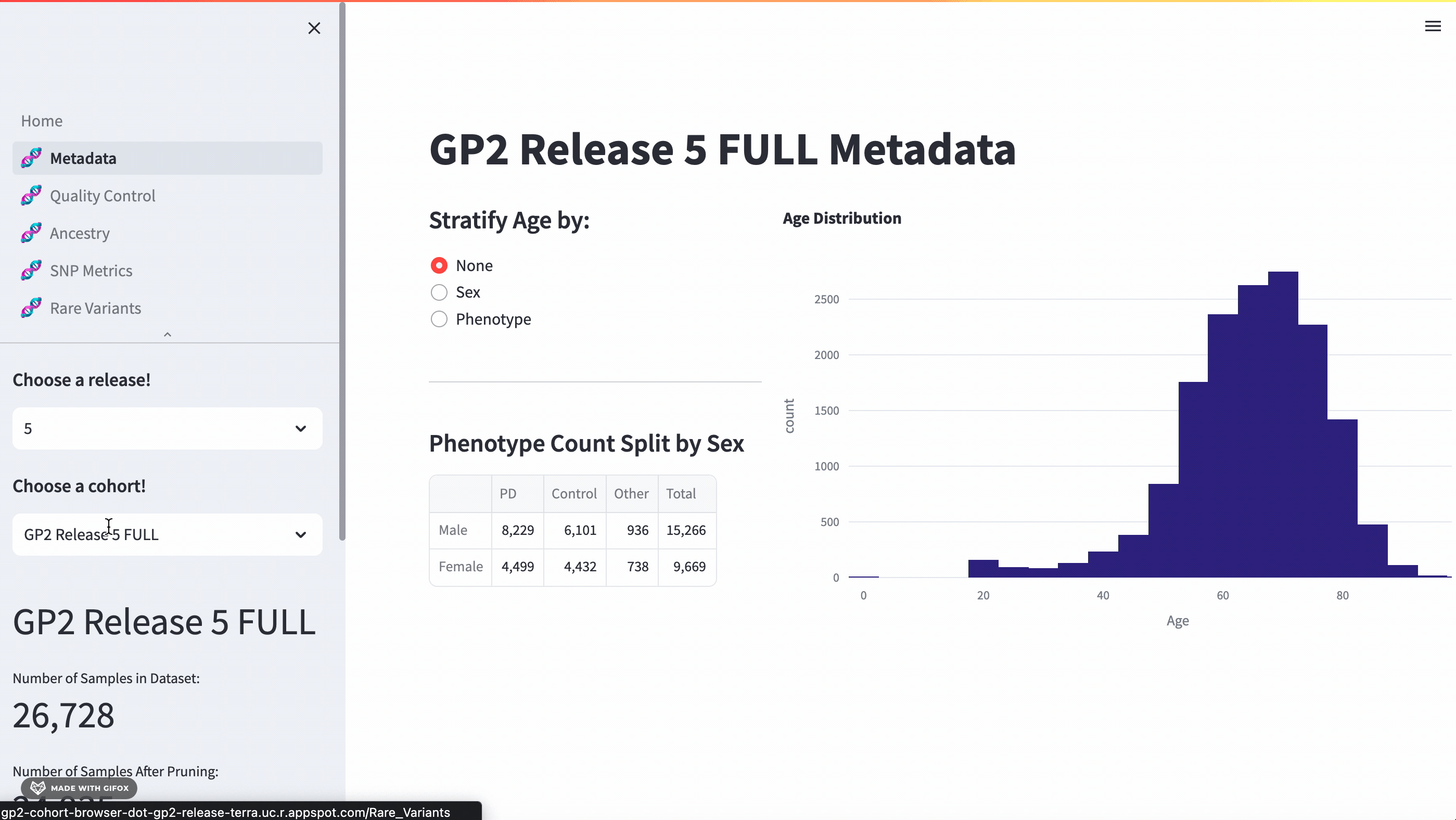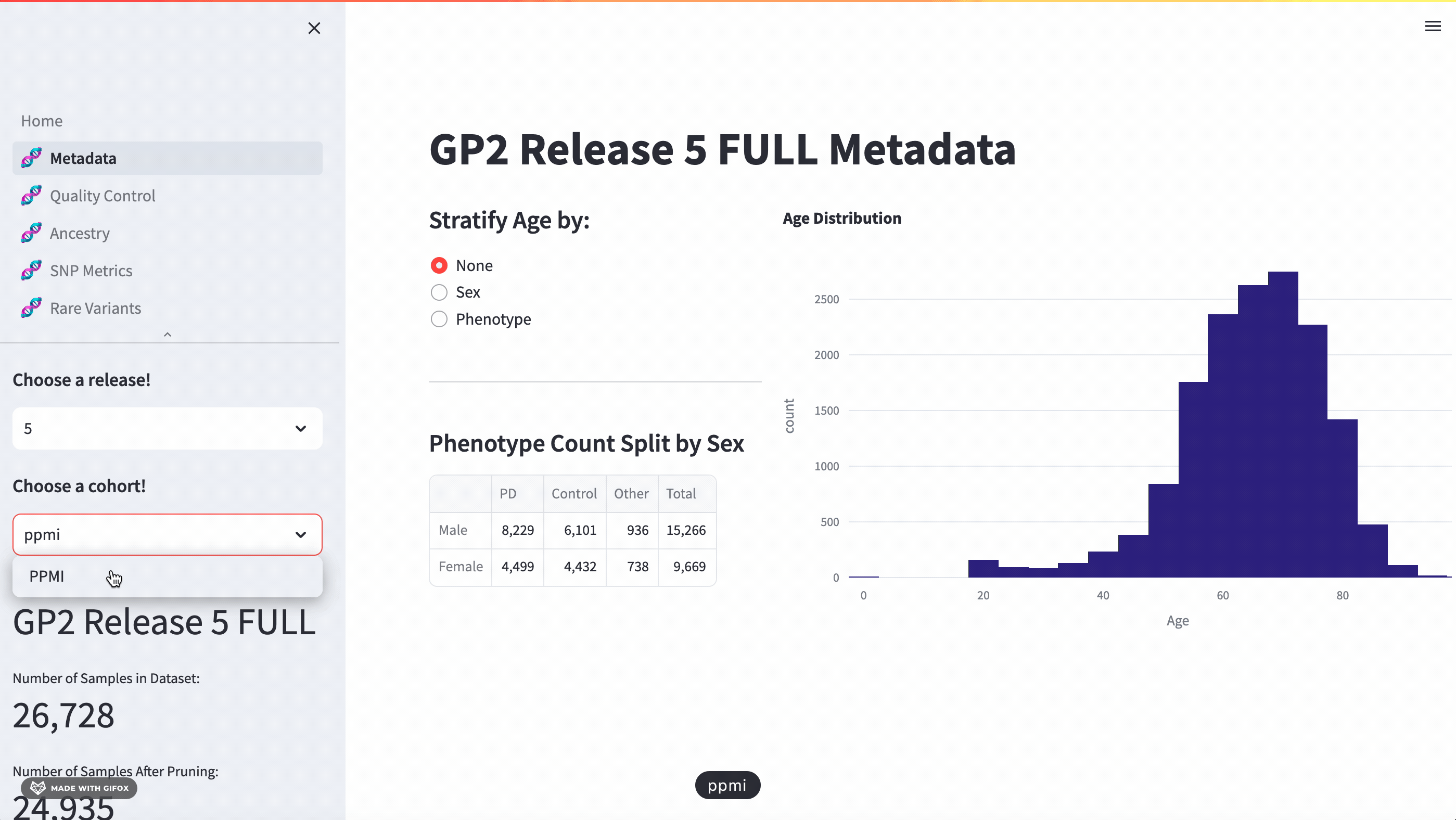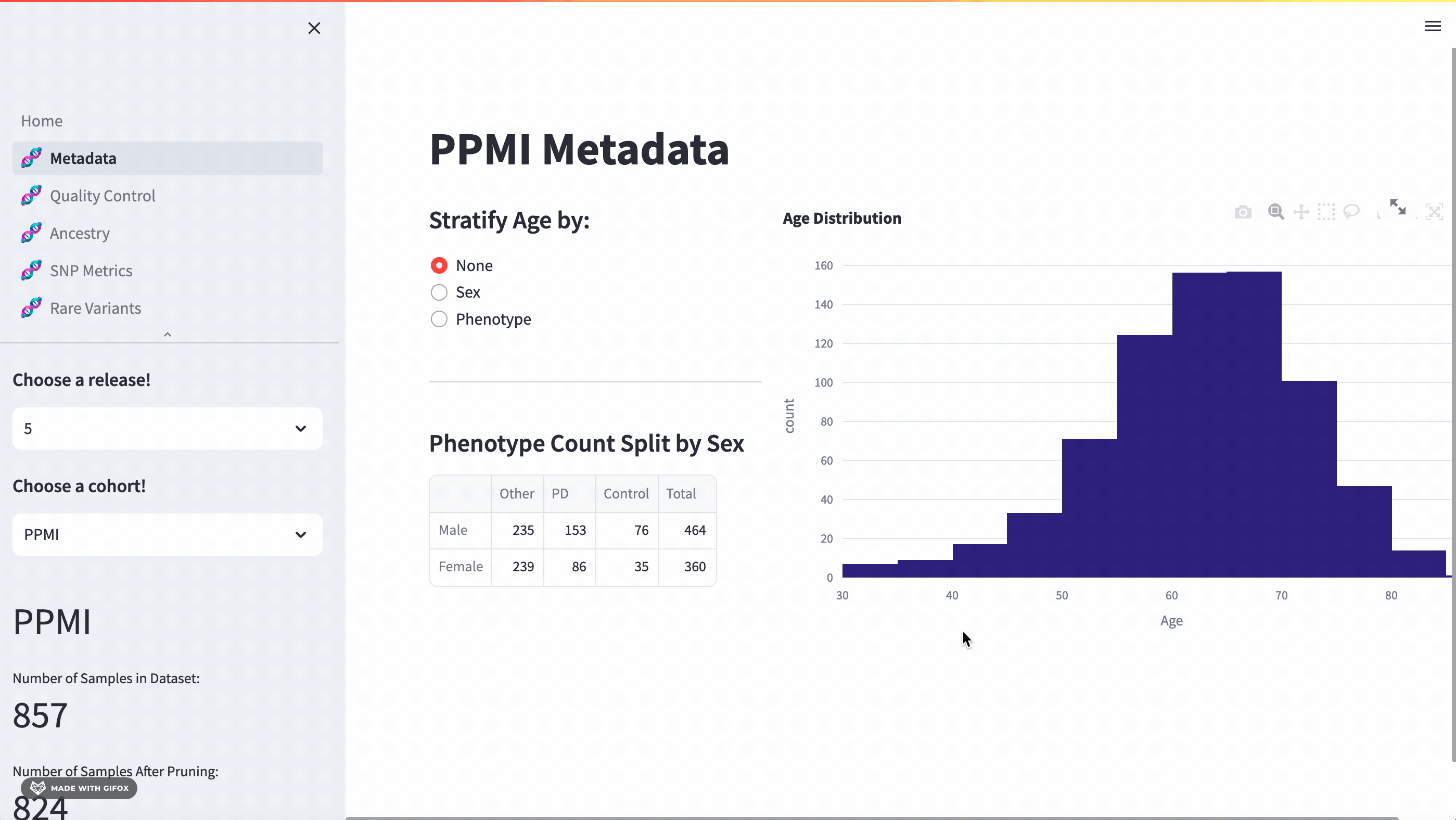
With this application, researchers can access a full meta-data breakdown, complete with histograms. It allows for detailed exploration of cohort demographics such as age, sex, and phenotype. Users can select which cohort they want to examine.
Quality Control
One of the key features of the GP2 Cohort Browser is its rigorous focus on quality control (QC). It allows you to access and understand our QC processes, including sample and variant level filtering. Moreover, you can also dive into the details of the updated QC description available in the dropdown section. The funnel plot visualizes the quality control process step by step, allowing you to get a better understanding of your data.
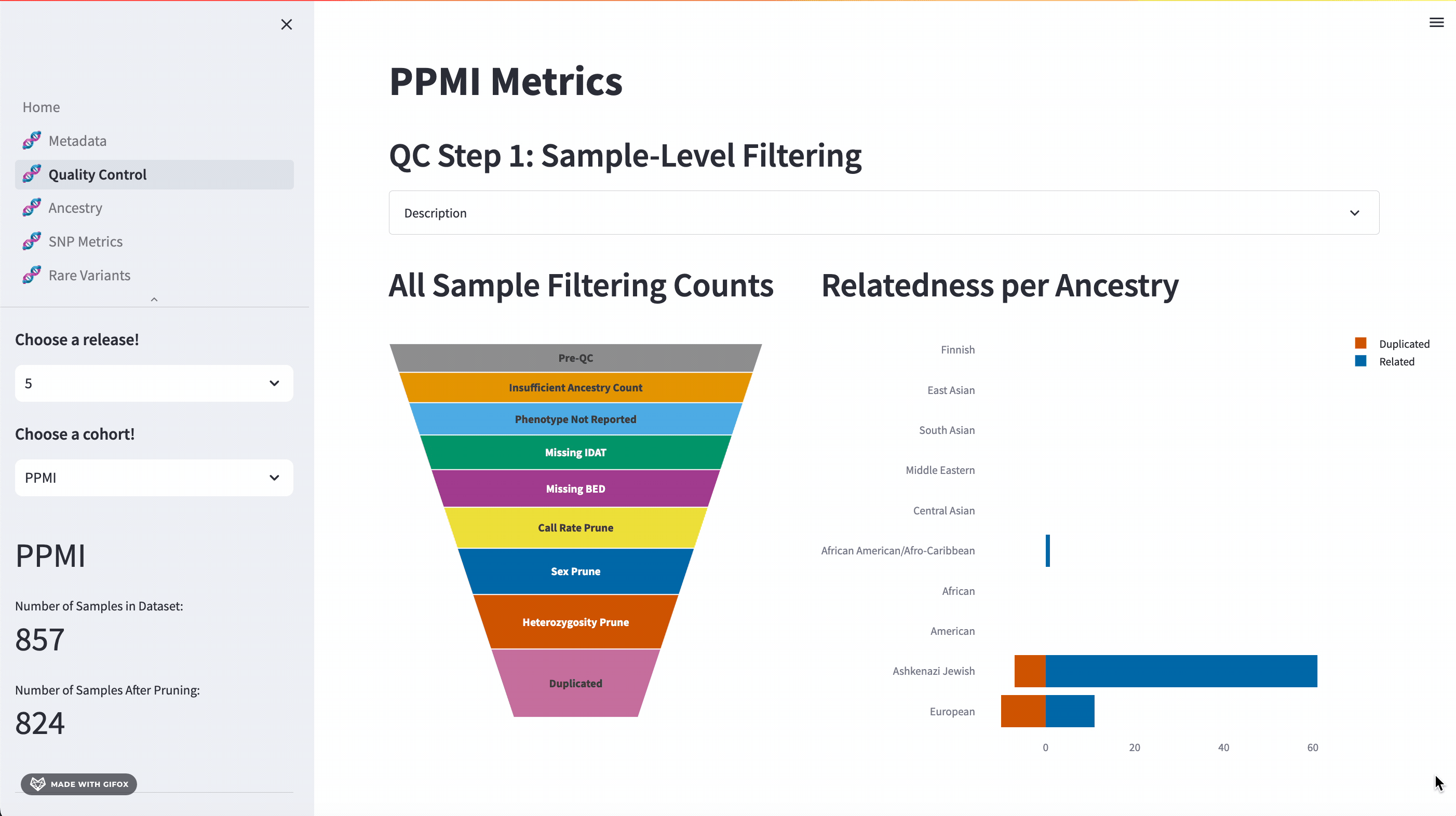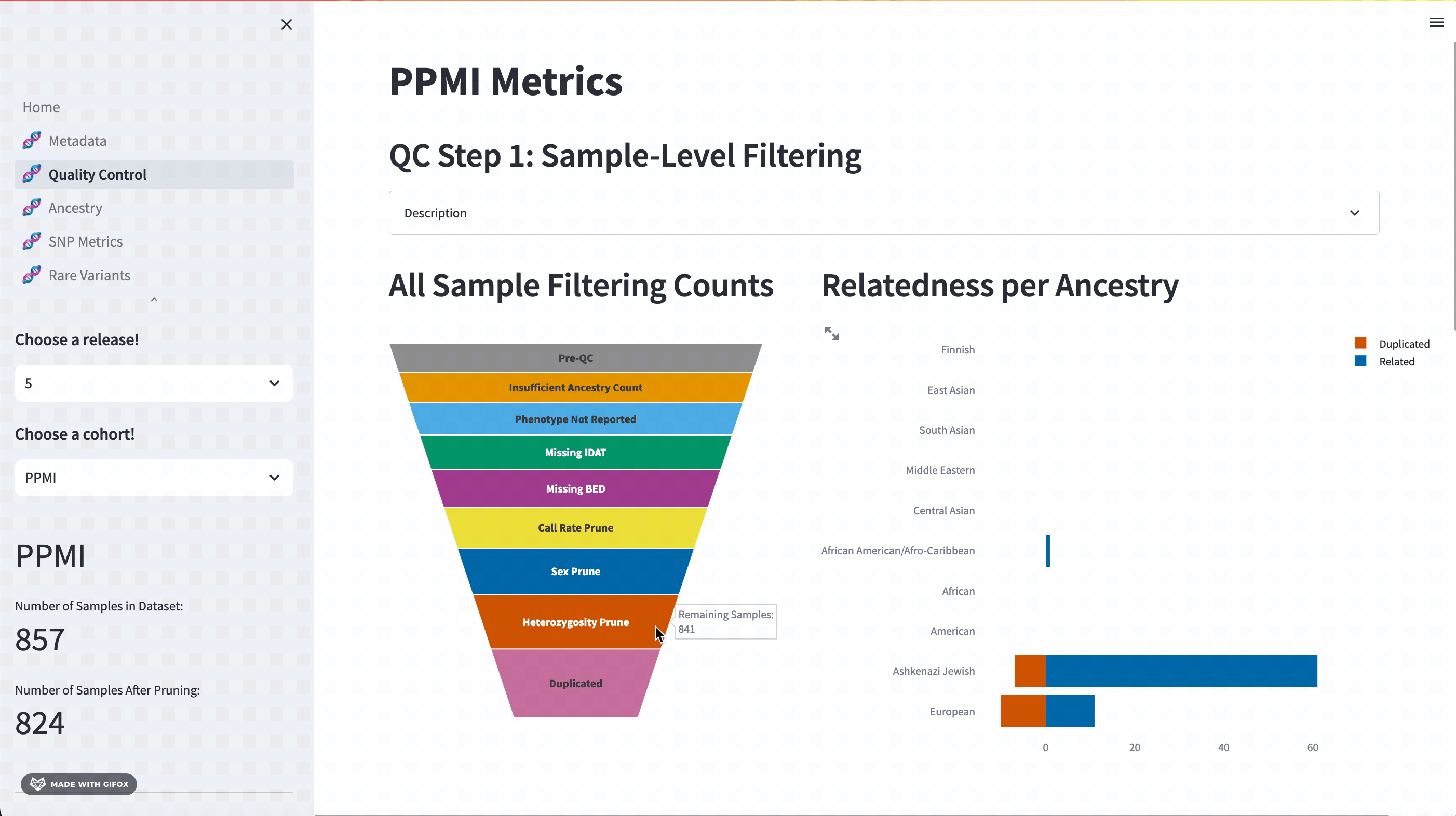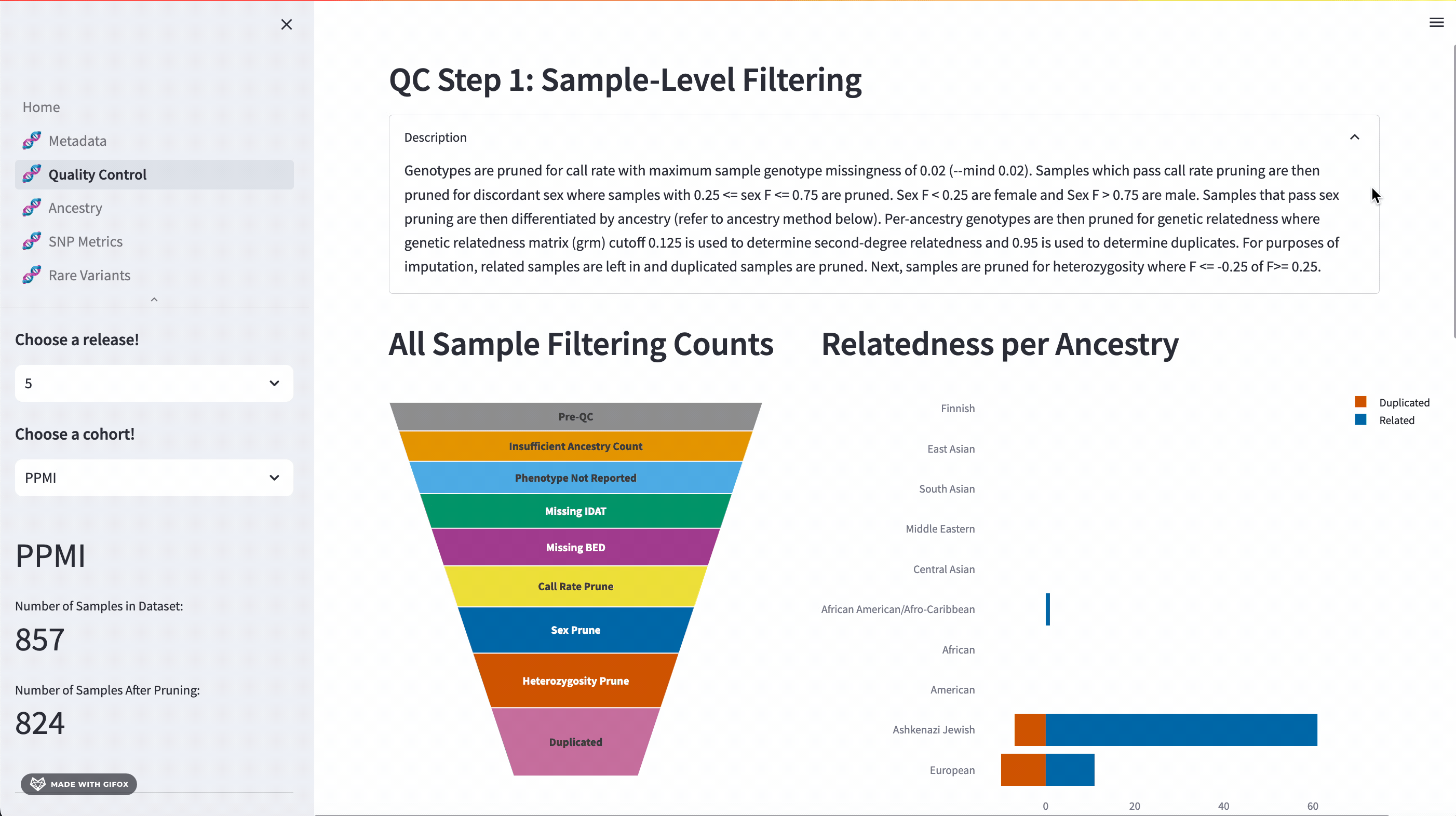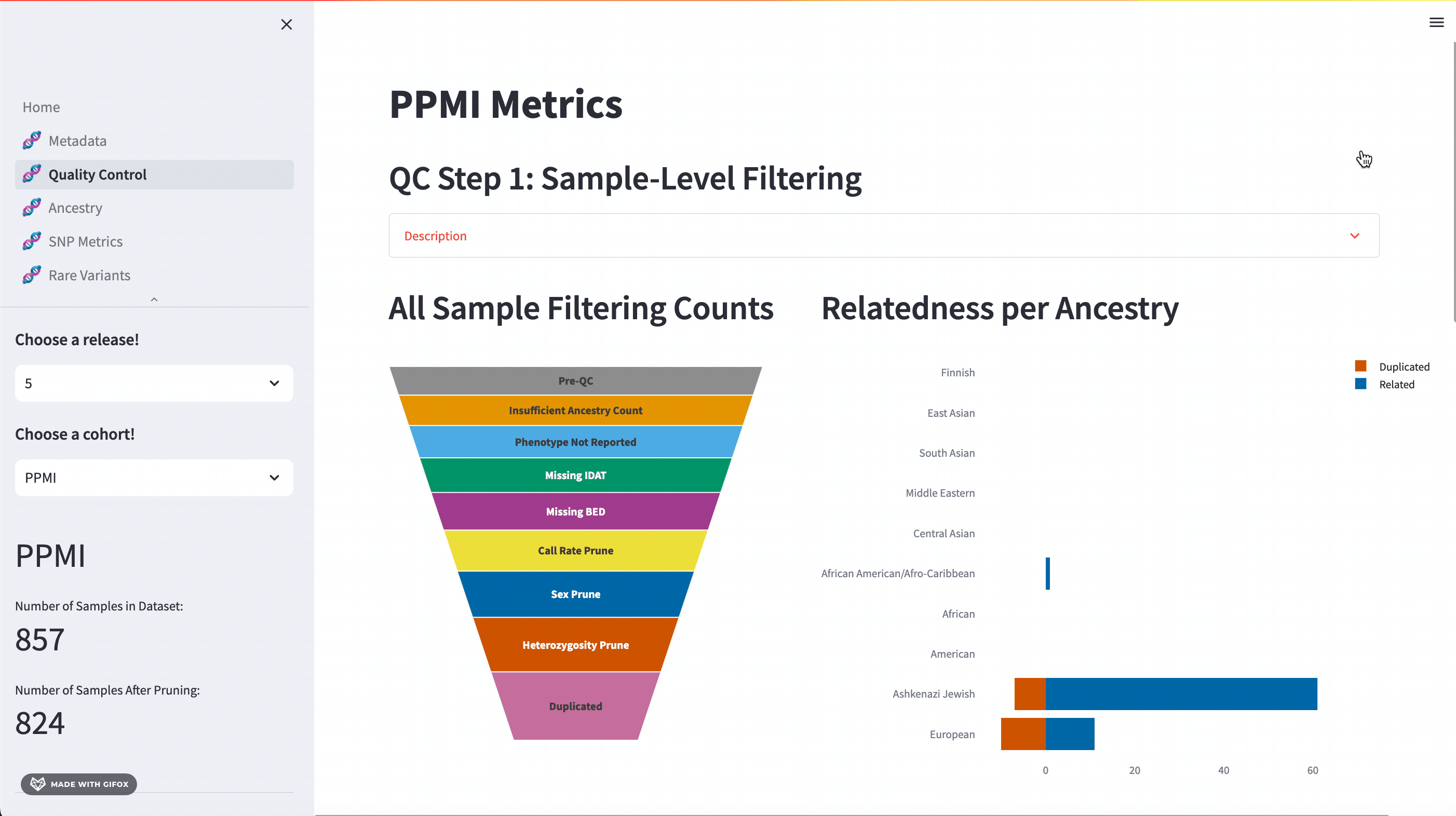
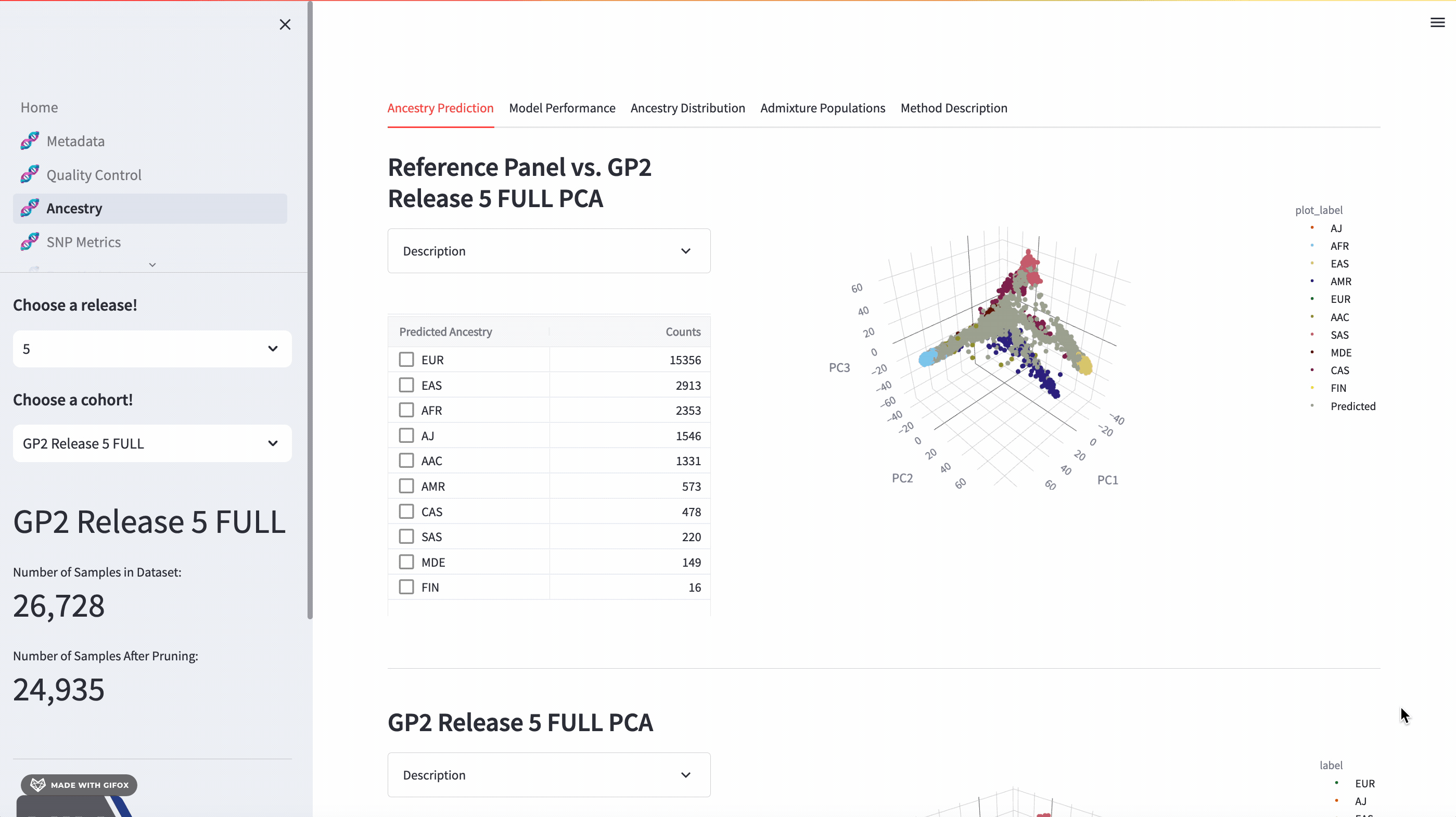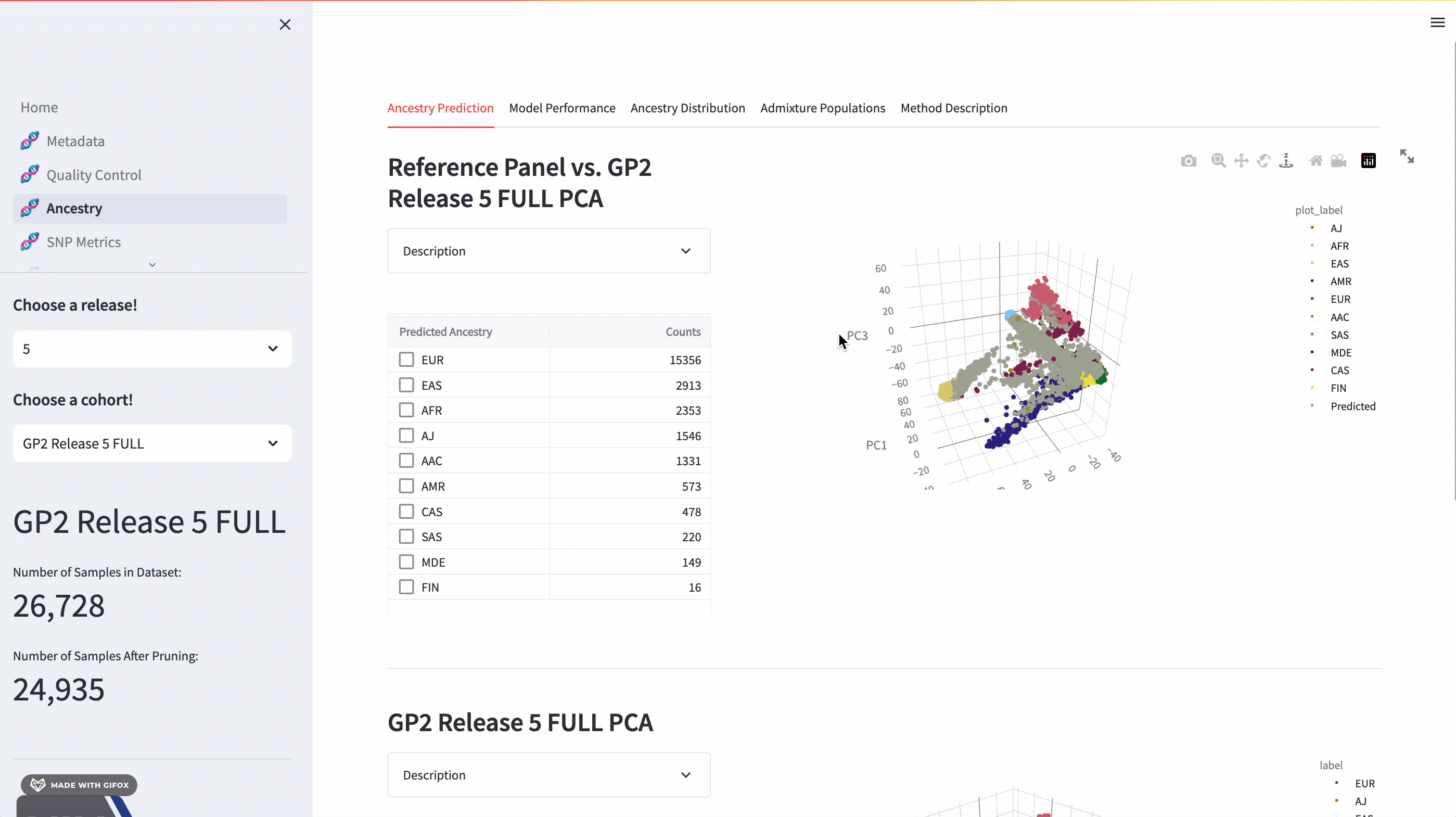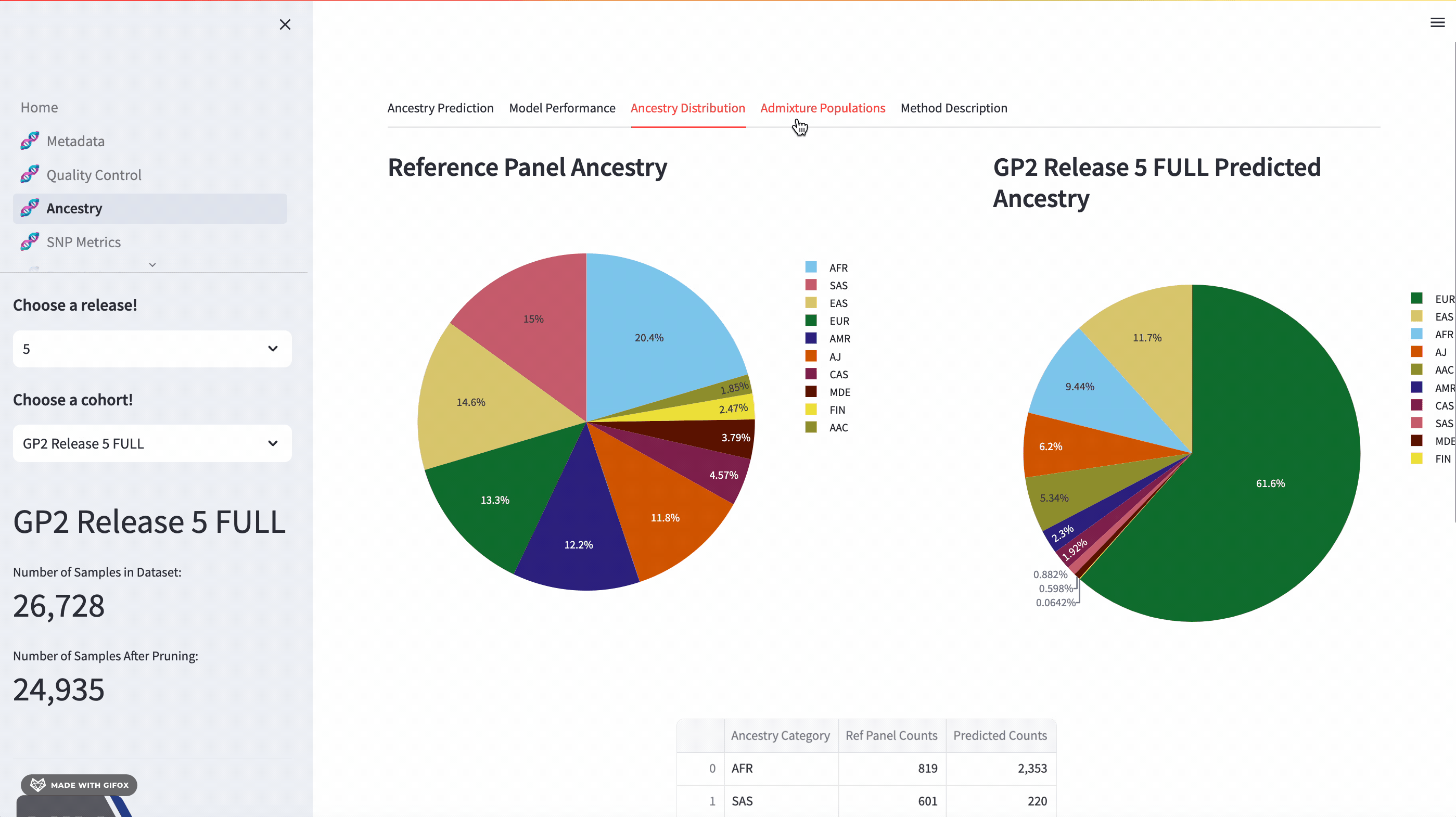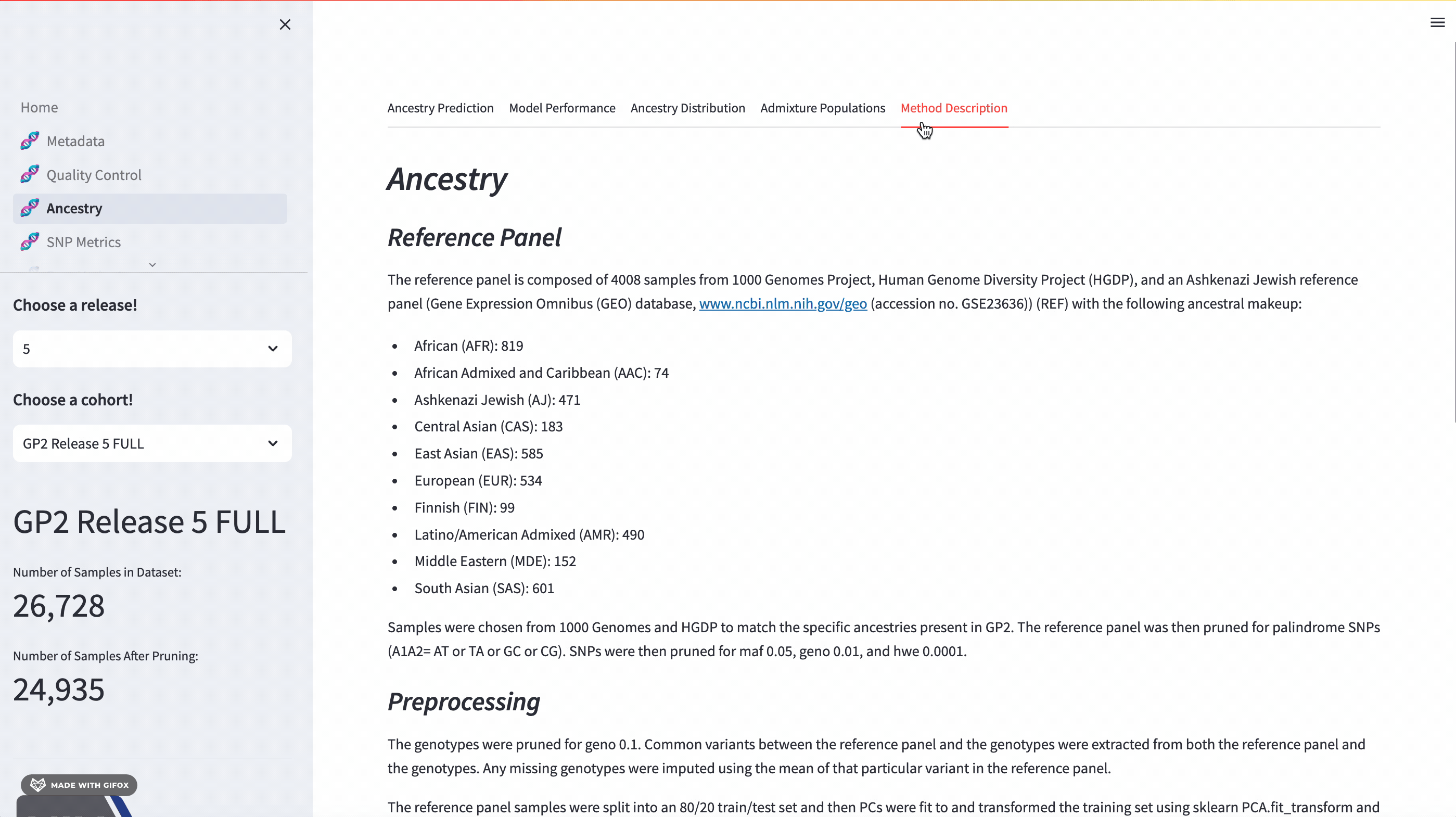
Ancestry Analysis
The GP2 Cohort Browser comes with a dynamic 3D Principal Component Analysis (PCA) plot that allows for an interactive exploration of genetic ancestry. The plot includes reference panels and lets you subset by predicted ancestry. Below the plot, you’ll find a table of predicted ancestry and a pie chart comparison to better understand the distribution. The application also presents model performance metrics such as the confusion matrix, accuracy, and F1 score.
Something to look forward to in the next release (expected release in early fall 2023) is expanding on the admixture feature for the reference panel, focusing on admixed populations such as Amerindian/Latino (AMR), Middle Eastern (MDE), and Central Asian (CAS) populations.
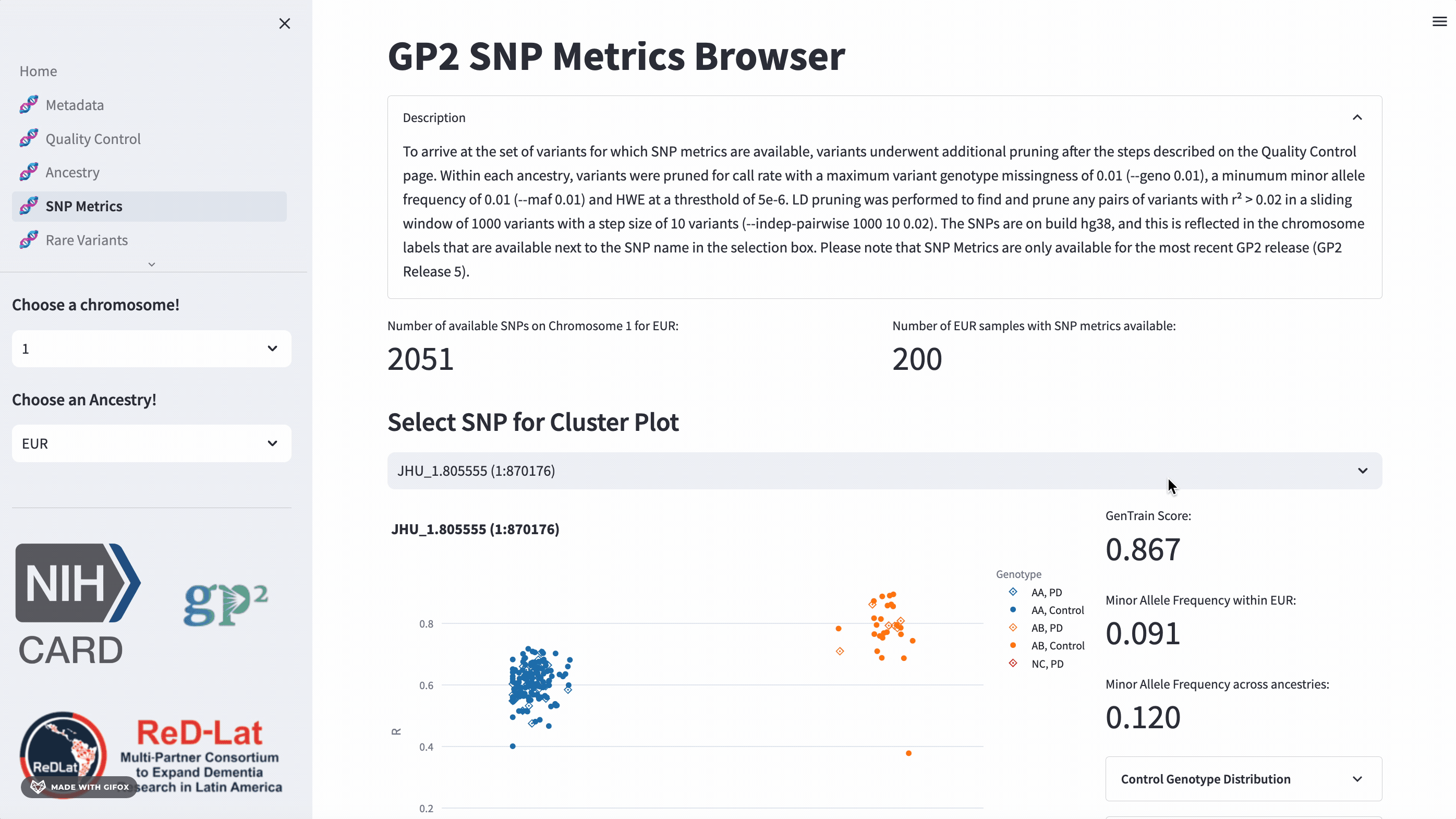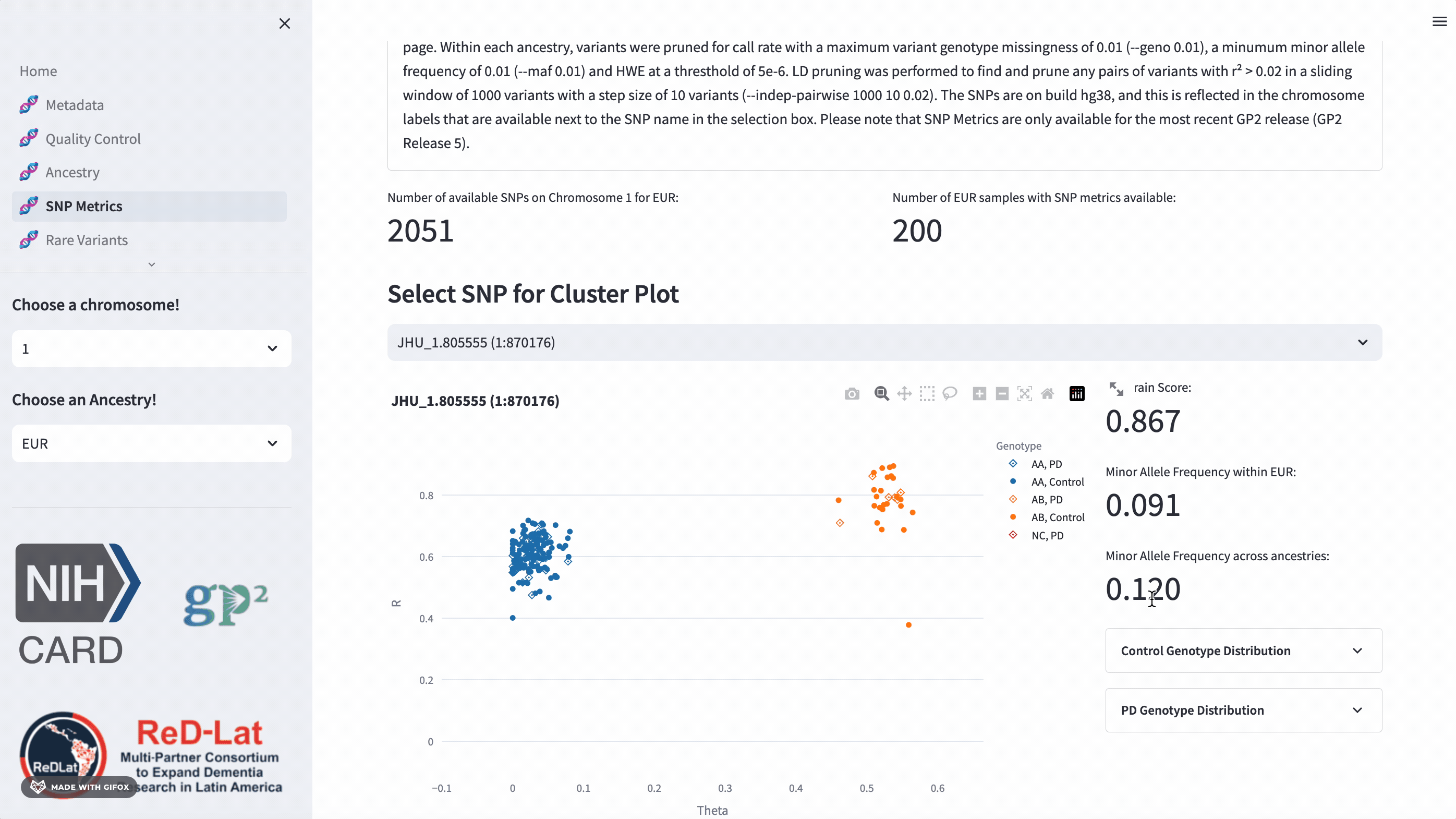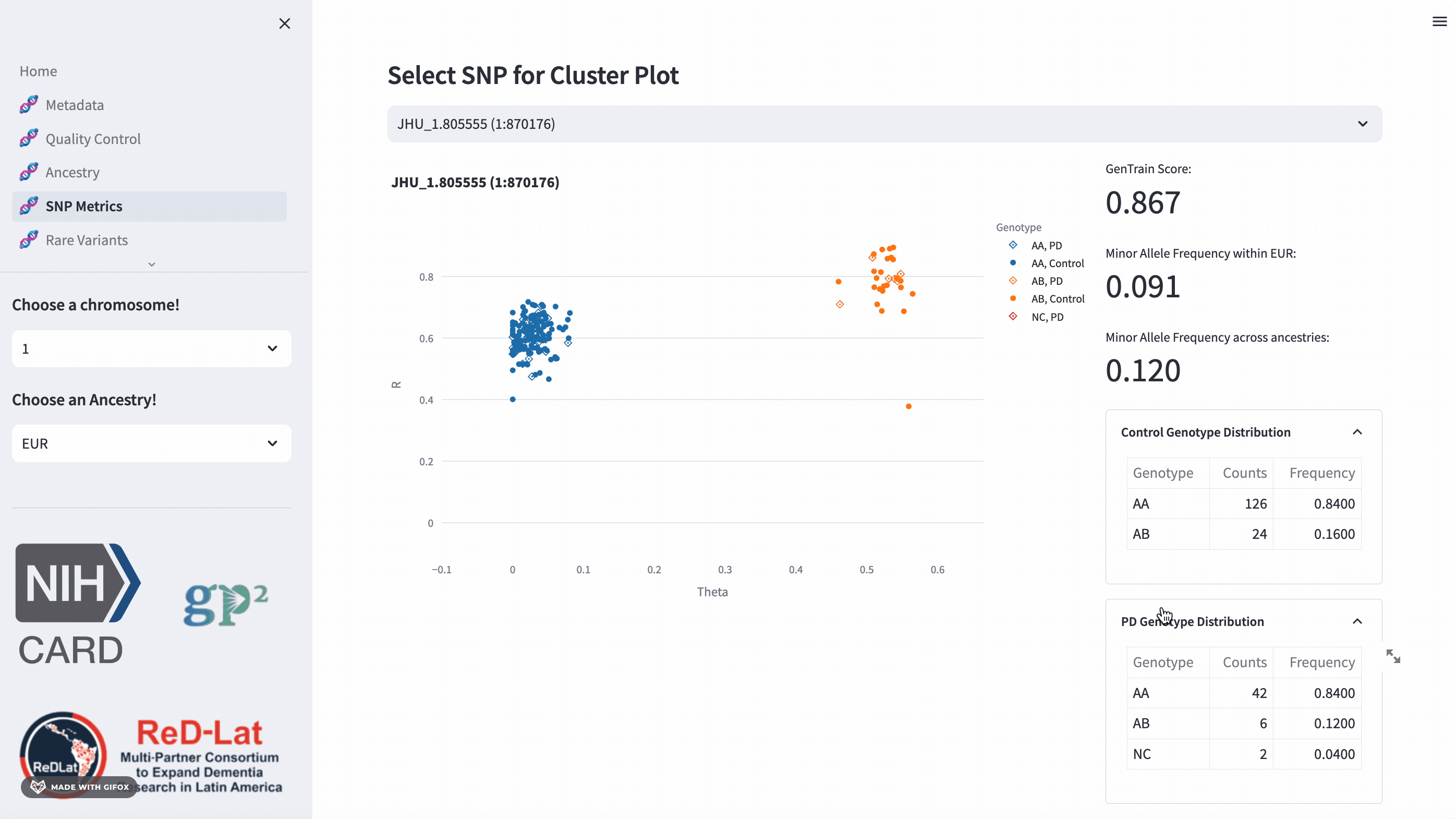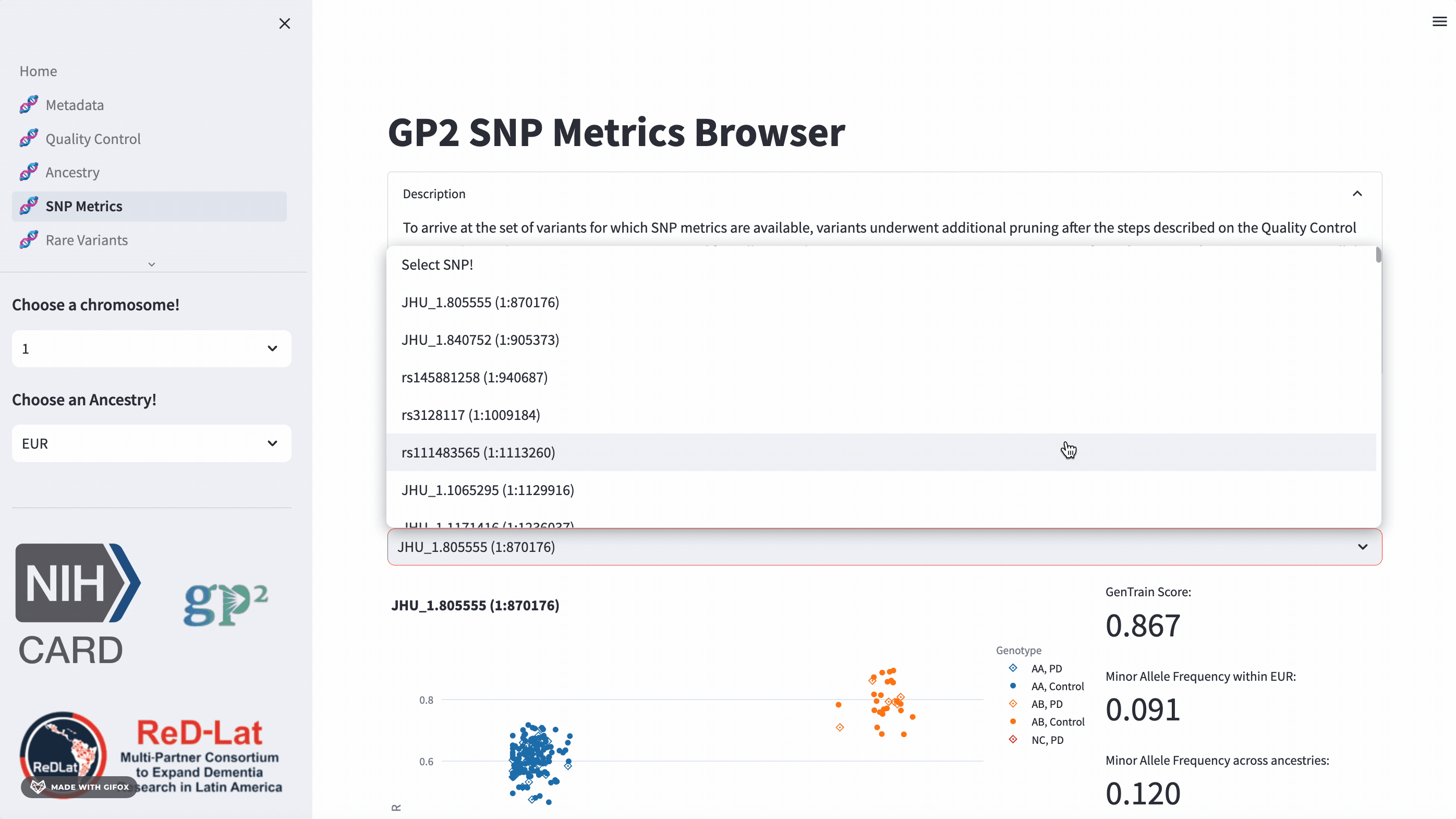
SNP Metrics
Researchers have the opportunity to explore single nucleotide polymorphisms (SNP) metrics for the most recent release, once they’ve selected an ancestry. Users can search the cluster plot by rsID or position (in hg38). This section also provides the GenTrain score and minor allele frequency distribution for control and disease groups.
Something to look forward to in our future releases is an expansion on the predicted clusters for lower resolution variants using our custom machine learning approach.
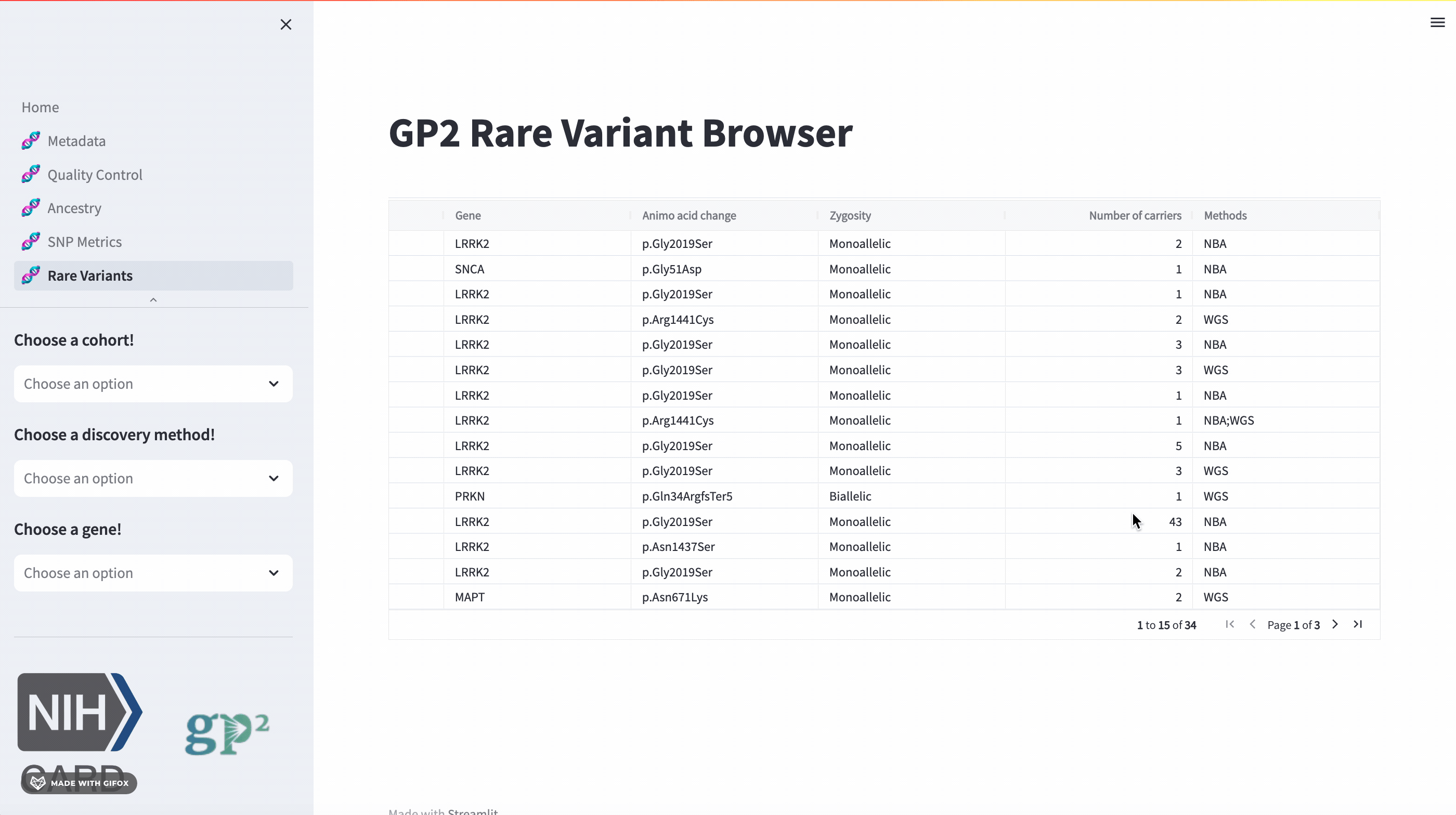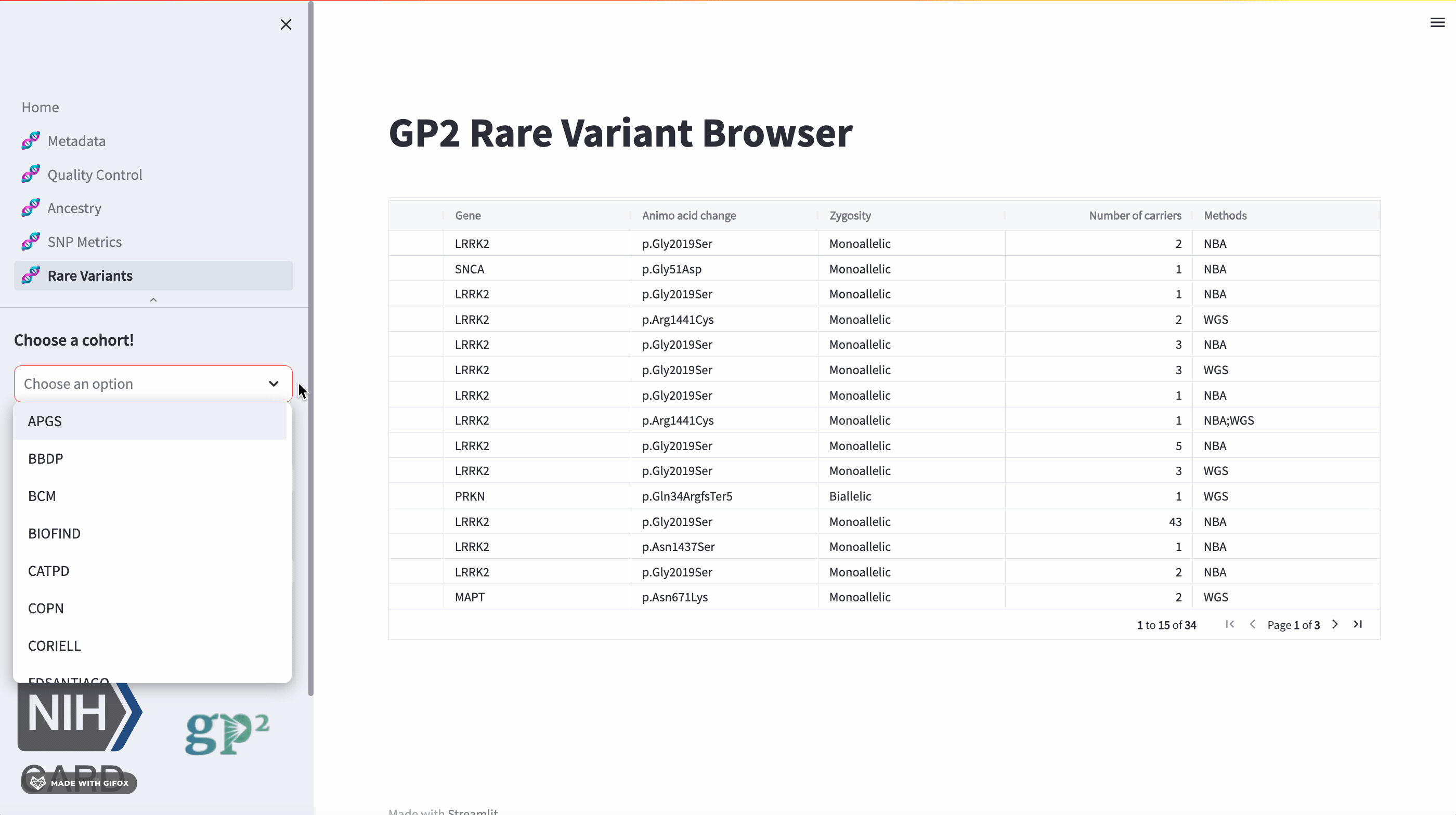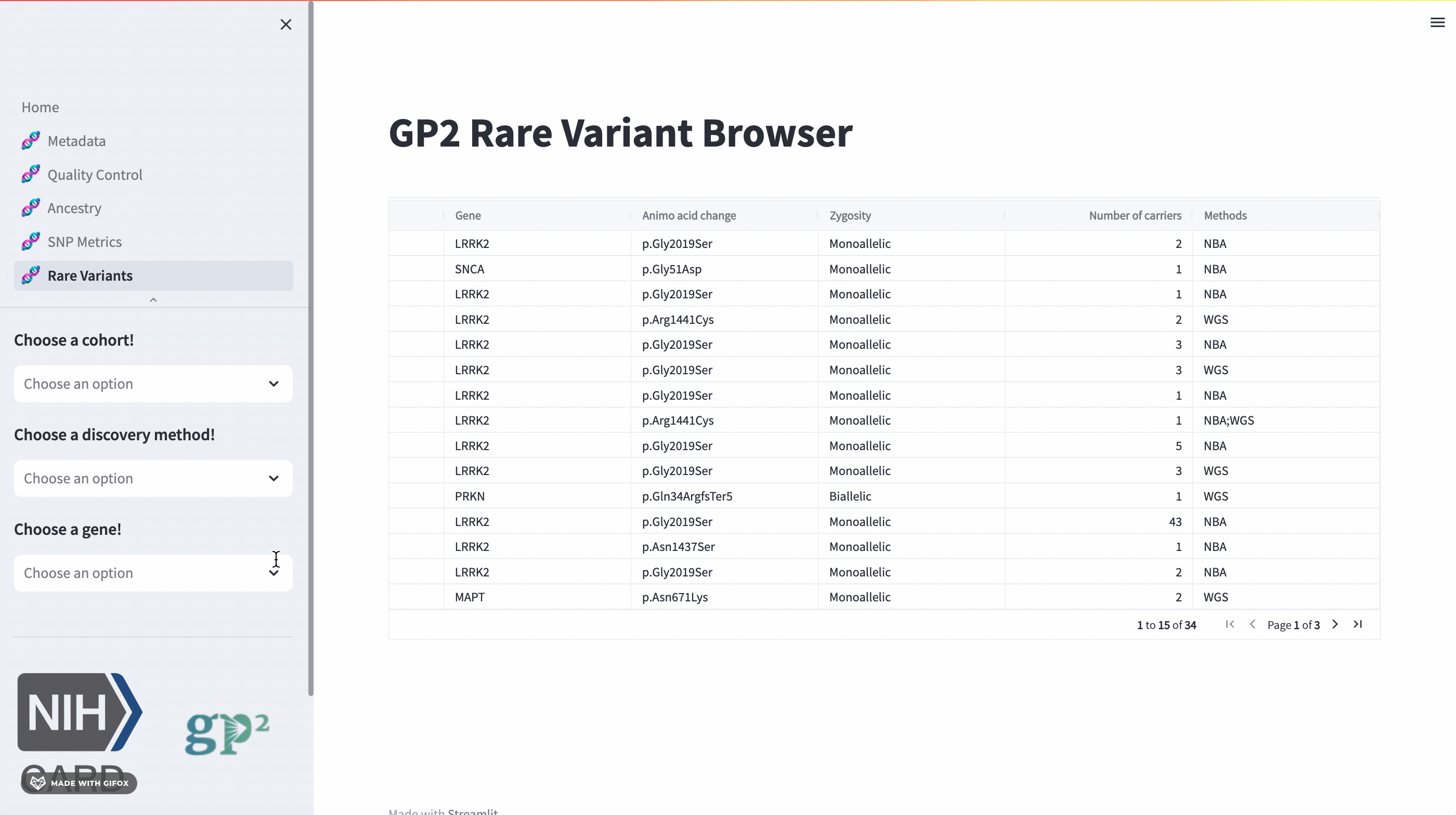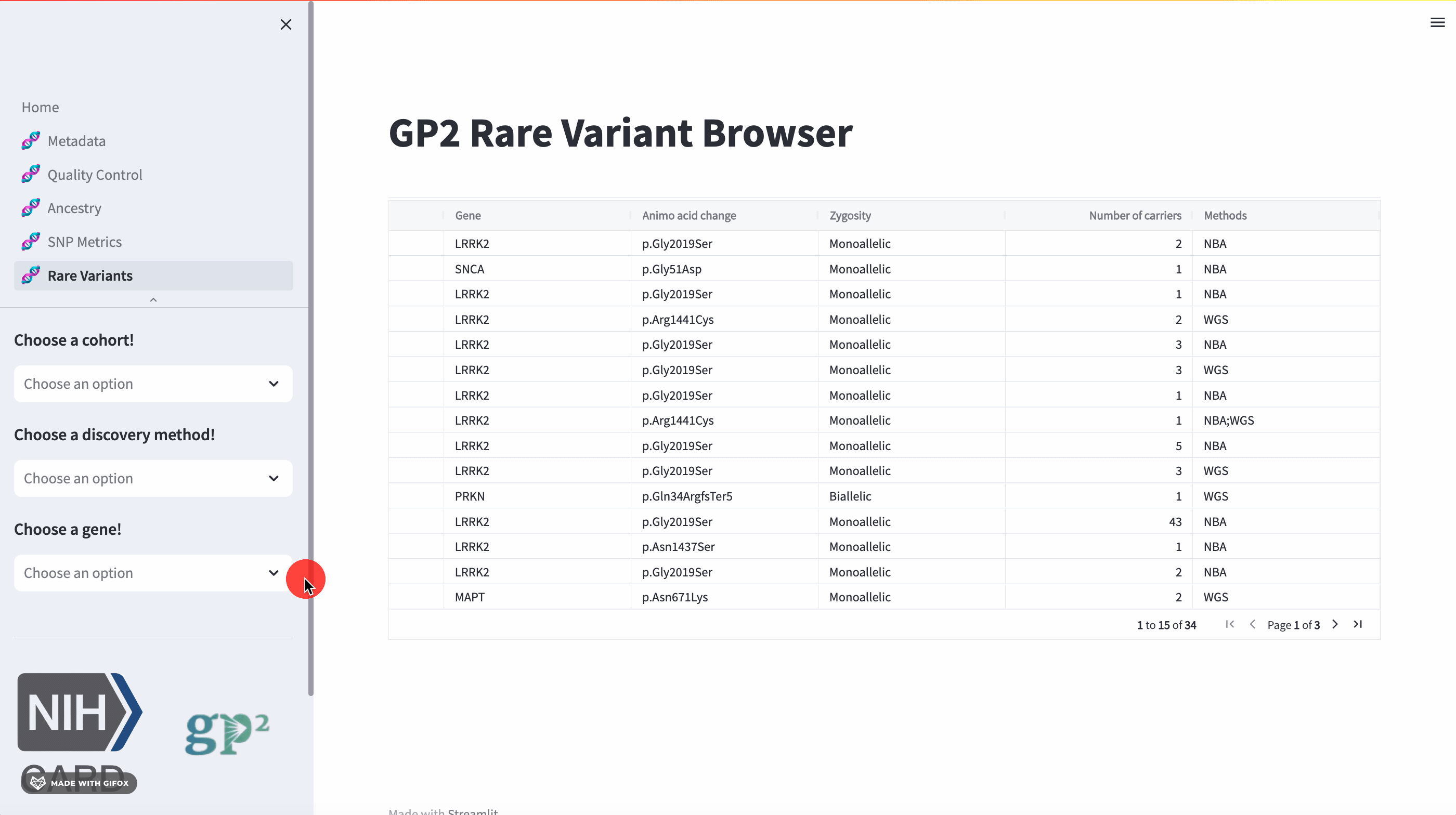
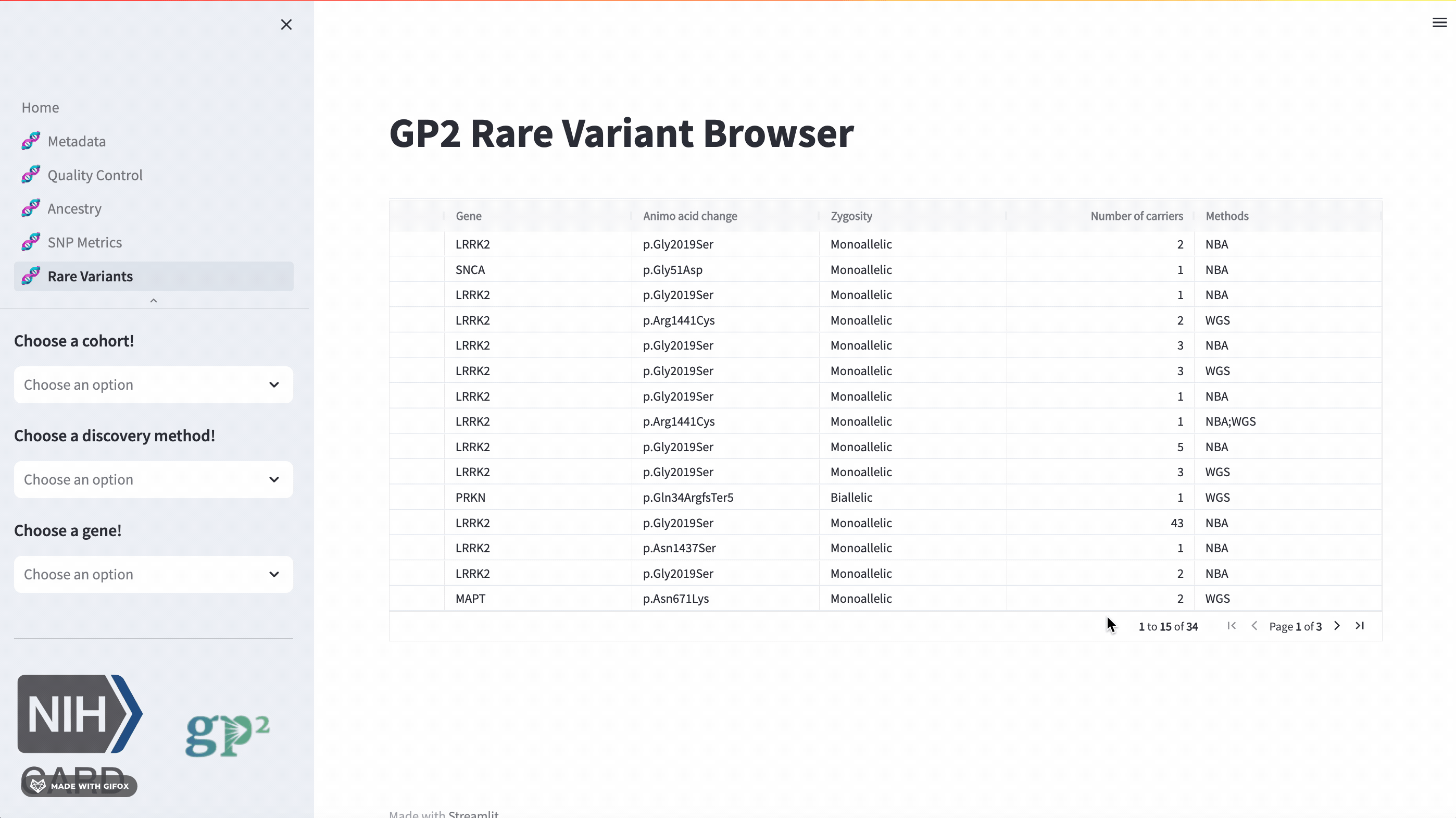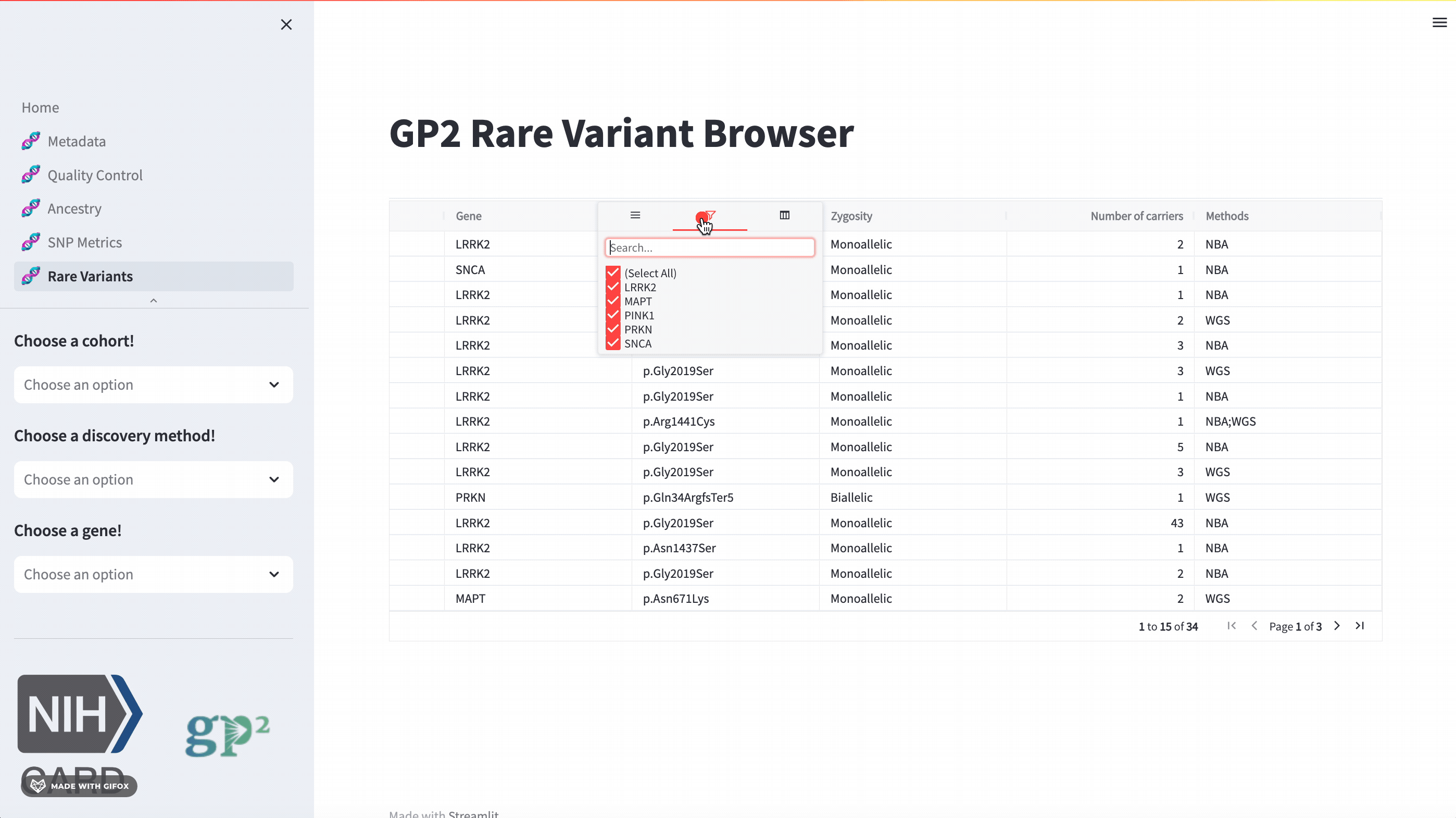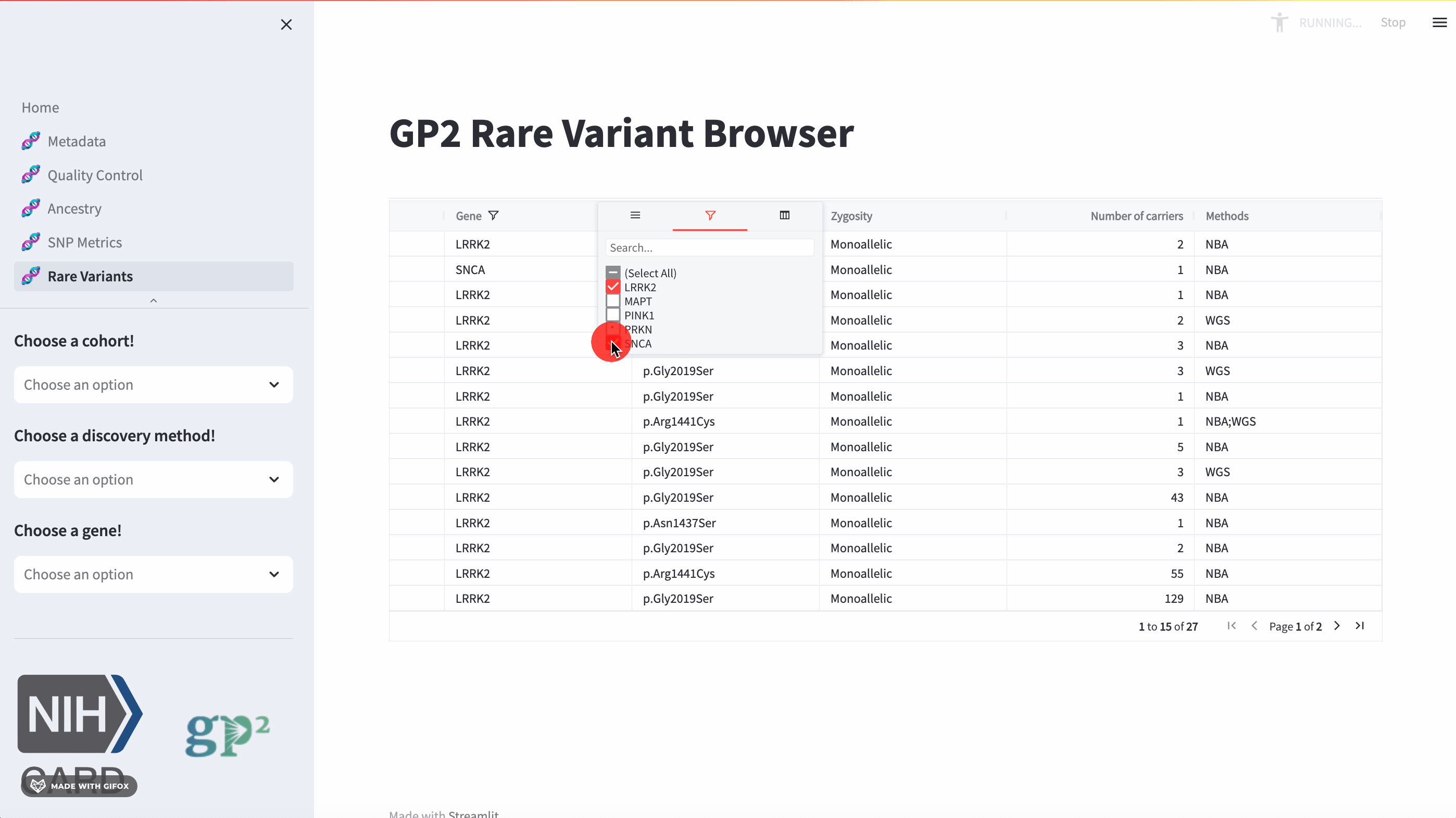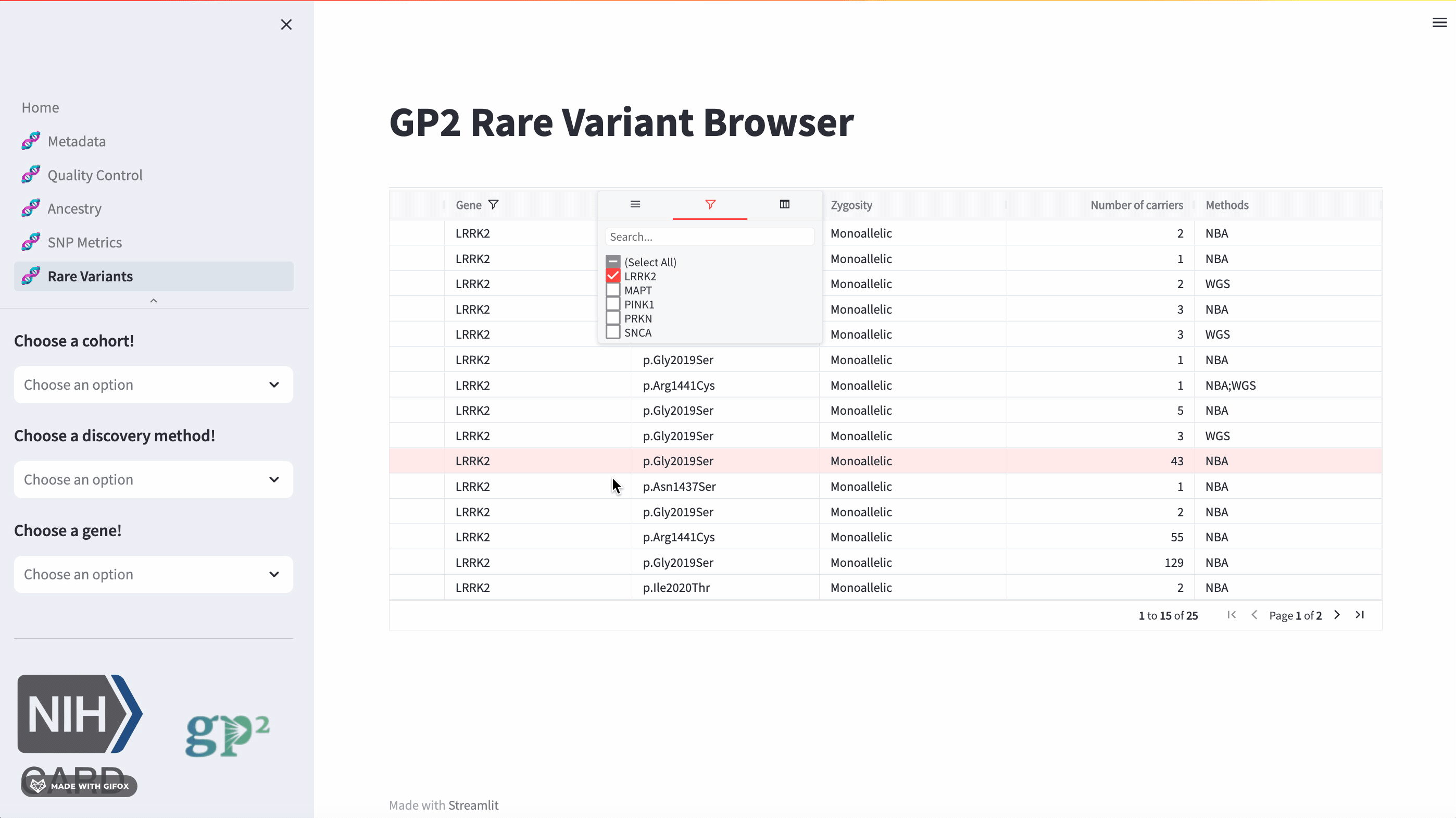
Rare Variants
The GP2 Cohort Browser also has a dedicated section for exploring rare variants. It includes information provided by GP2’s Monogenic Network where known PD genes can be subset to be investigated closer. The sidebar filter allows users to search for specific genes, while the table provides comprehensive data on all other aspects.
Final Thoughts
We sincerely hope the GP2 Cohort Browser is a tool you can use to better understand data available as a part of GP2 and for creating summary figures for papers and posters.
The GP2 Cohort Browser, which is consistent across releases and cohorts, is a bold step forward increasing transparency and accessibility regarding each step of the QC process within GP2. We thank all of the cohorts around the globe who have donated data for the GP2 effort and make this entire undertaking possible.
In case of any queries or support, please contact the Data Analysis Working Group [email protected].





