The Global Parkinson’s Genetics Program (GP2) and Accelerating Medicines Partnership: Parkinson’s Disease (AMP® PD) have partnered to be your one-stop shop for Parkinson’s disease genetic (GP2) and genomic (AMP PD) data that will speed your discovery of novel drug targets and biomarkers to accelerate generation of new medicines for Parkinson’s. We are adding collaborative cloud-based computing to connect these two massive databases.
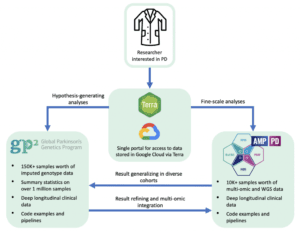
We are providing a single safe and easy access point to a massive amount of data stored in the cloud, using Terra as the backbone for data analysis, data storage, and researcher authentication. Terra takes computational work to the next level, with collaborative workspaces containing cloud native notebooks and workflows. Cloud-based computing means you can analyze the data in a more secure place without time-consuming and expensive data transfers. Terra’s use of intuitive Jupyter notebook structure allows for clear and interactive analysis, and you can also publish your workspaces along with your results to ensure reproducible and transparent research. Future plans include interoperability with other neurodegenerative disease workspaces and platforms to further maximize the data’s value.
Why place these two programs in one place? Because they are nicely complementary. If you want a deep dive into ten thousand samples with various harmonized multi-omic read-outs, including RNAseq and genomeSeq, then AMP PD is your dataset. If you want to explore genetic associations in diverse populations on a biobank scale, head to GP2. Both GP2 and AMP PD also include curated relevant longitudinal clinical data on most participants. In general, we use GP2 as a discovery dataset for massive scale analyses (hypothesis generating), and then potentially further refine and closely explore these discoveries in the AMP PD unified cohorts.
The best part is, the same access point and paperwork allow access to both datasets, with a single architecture and extensive documentation available to reduce the learning curve. We developed this graphical overview to illustrate the use of cases for GP2 and AMP PD data.
This blog was written by the following members of GP2 and AMP PD:
Mike A. Nalls, PhD
Data Tecnica International | USA
Mike founded Data Tecnica in early 2017 after over a decade of experience in large dataset analytics and methods research in healthcare and other scientific fields. 350+ peer-reviewed publications (before the age of 40) in the field of applied statistics in large datasets, brain diseases, and genomics. He is a strong advocate of open science, collaboration, and transparency in science.
Hampton Leonard, MS
Data Tecnica International / National Institutes of Health | USA
Hampton has a background in data science and machine learning, which she applies to large multi-omic datasets in the neurodegenerative disease space. She is passionate about investigating differences on both clinical and omic levels and how these differences can affect clinical trial outcomes.
Matt Bookman
Verily Life Sciences | USA
Matt is a Solutions Architect for the Terra team at Verily. As co-chair of the AMP PD Data Working Group, he worked with collaborating scientists to process genomic and transcriptomic data and make it available to qualified researchers in Terra. Matt received a BS in applied mathematics at UCLA and an MS in computer science and computational biology at Stanford University.
Eline Appelmans
Foundation for the National Institutes of Health | USA
As Director of Neuroscience Research Partnerships, Dr. Appelmans works in coordination with the NIH, non-profit, industry leaders, and FNIH project leads on the teams’ implementation of the Accelerating Medicines Partnership (AMP) for Alzheimer’s Disease (AMP-AD), AMP for Parkinson’s Disease (AMP PD), AMP for Schizophrenia (AMP-SCZ), the Alzheimer’s Disease Neuroimaging Initiative 3 Private Partner Scientific Board (ADNI3 PPSB) and the Biomarkers Consortium Neuroscience Steering Committee (BC NSC), and other projects in development.





