Parkinson’s disease (PD) has been the focus of extensive genetic research. Researchers have identified over 90 genetic variants that may affect a person’s risk of developing PD, revealing that this is a complex disease involving many molecular pathways.
However, a big-picture, global understanding of PD has been hampered by a lack of genetic diversity in PD studies, which have largely focused on European ancestry populations. Which PD genetic variants are more likely causal? Which variants might be clinically useful across populations? Answering these questions requires more ancestrally diverse data.
A Global Collaboration: Diverse Ancestry and Impressive Scale
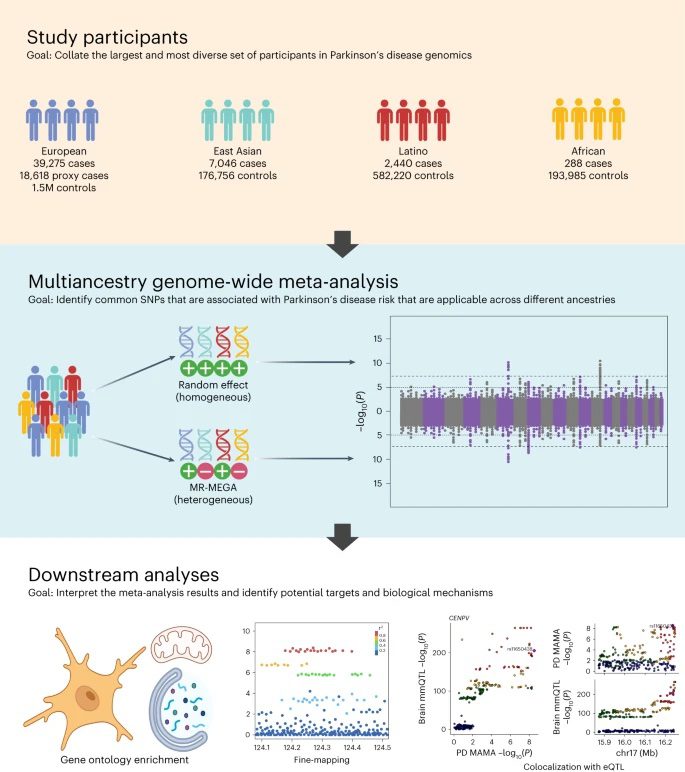
A new collaborative study published in Nature Genetics offers a more inclusive and global understanding of PD’s genetic landscape. The study is the largest and most diverse of its kind, thanks in large part to global collaboration and data-sharing efforts spearheaded by the Global Parkinson’s Genetics Program (GP2). This study analyzes data representing 49,049 individuals with PD, 18,785 individuals whose parents have or had PD, and over 2 million neurologically healthy individuals of European, East Asian, Latin American, and African ancestry.
Researchers from NIH, GP2, 23andMe, and collaborators in the US, UK, Peru, and Singapore curated this ancestrally diverse data from FinnGen, 23andMe, UK Biobank, and other open-access data repositories. With this data, they identified 78 regions of the genome (“loci”) significantly associated with PD.
Analyzing data from multiple different ancestry groups has unique challenges, but new software and analysis tools are emerging that take advantage of increasingly diverse data. Thanks to these tools and collaboration efforts across the globe, researchers can now identify genetic regions significantly associated with PD risk across multiple ancestral populations. Such regions are more likely to contain causal variants.
For the study, researchers used an approach called a multi-ancestry genome-wide association (GWAS) meta-analysis. GWAS is a method that involves surveying the genomes of many people, looking for specific sequence variants in genomic regions (“loci”) that occur more frequently in those with a specific disease compared to those without the disease. The researchers pulled together data from previously published GWAS and diverse data repositories to add statistical power and robustness to the search for PD genetic risk factors.
By including data from non-European populations, the researchers were able to identify many novel genetic loci that hadn’t before been observed when only looking at European-derived populations. While a majority of the 78 PD loci identified in this study match up with regions already associated with PD, 12 of them are potentially novel. The novel loci appear to involve genes that help break down or metabolize proteins, control the activity of mitochondria, and carry out immune responses in the brain.
Most of the identified loci were significantly associated with PD risk across all of the ancestries studied, including European populations. These results suggest that the many pathways that may cause PD are shared across populations.
Towards Personalized Medicine: Fine-Mapping and Functional Implications
The researchers were also able to pinpoint the specific causal genetic variant within at least 6 of the loci they identified, in a process called fine-mapping. Fine-mapping estimates which specific variants within each loci are likely responsible for conferring disease risk in a certain population.
“One of the issues with GWAS is that you never know which specific variant is the functional or causal one at an identified locus,” said Ignacio ”Nacho” Mata, a scientist at the Learner Research Institute, assistant professor of molecular medicine at the Cleveland Clinic Learner College of Medicine, and an author of the study. “At any given GWAS ‘hit’ or region of the genome significantly associated with disease, there might be hundreds of potential candidate variants.”
Identifying the functional variants at significant GWAS loci and how they affect gene expression is key to follow-up studies and the development of targeted therapeutics for PD.
“By incorporating ancestrally diverse datasets, we were able to narrow down the functional variants for some of the genes at the loci we identified,” Nacho said. “Ancestrally diverse data allows you to identify variants that might be population specific, for example, and also allows you to identify variants that are significant across different populations and thus more likely to be important causal risk factors.”
Some of the identified loci did have different effects between different populations, highlighting the importance of genetic studies that dive into ancestry-specific risk factors. Two loci found only in Latin American and African populations were near the JAK1 and HS1BP3 genes. These genes are both involved in inflammatory signaling and may support the role of inflammation in PD.
Implications and Future Directions
The study’s findings emphasize the importance of including diverse ancestries in genetic research. This inclusivity enhances the accuracy of genetic risk prediction and the development of personalized treatments. GP2 is partnering with institutions that work with underrepresented populations to generate data for these underserved communities all over the world.
“We had people from all around the world working together and learning from each other,” Nacho said. “To me, this collaboration was the most exciting thing about this study and future possibilities.”
Further lab studies will be needed to confirm how specific loci identified in this study affect PD pathways and disease manifestations. However, this work lays a robust foundation for future studies to build upon.
This work was supported by: Intramural Research Program of the National Institutes of Health (NIH), National Institute on Aging (NIA), NIH, Department of Health and Human Services; National Institute of Neurological Disorders and Stroke; Parkinson’s Foundation; Michael J Fox Foundation; Aligning Science Across Parkinson’s Global Parkinson’s Genetic Project (ASAP-GP2); American Parkinson’s Disease Association; National Medical Research Council Singapore; and Singapore Ministry of Education Academic Research Fund.
This research has been conducted using the UK Biobank Resource under Application Number 33601. We want to acknowledge the participants and investigators of the FinnGen study. We thank the research participants and employees of 23andMe. Data used in the preparation of this article were obtained from Global Parkinson’s Genetics Program (GP2). GP2 is funded by the Aligning Science Against Parkinson’s (ASAP) initiative and implemented by the Michael J. Fox Foundation for Parkinson’s Research (https://gp2.org).





