Overview
In July 2025, GP2 announced the 10th data release on the Terra and the Verily® Workbench platforms in collaboration with AMP® PD. This release includes 11,109 additional genotyped participants and 13,339 additional WGS participants.
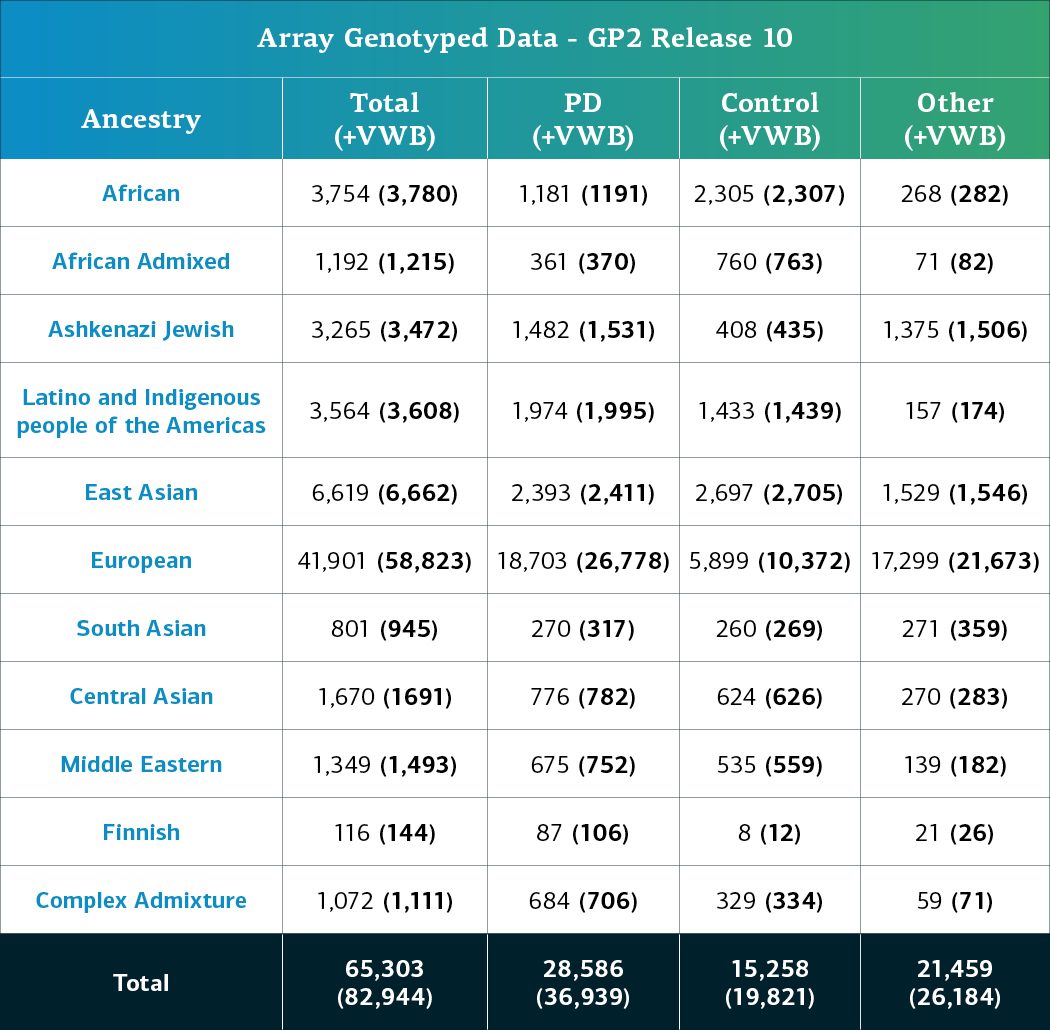
- The genotype array (NBA) data, including locally-restricted samples, now consists of a total of 82,944 genotyped participants (36,939 PD cases, 19,821 Controls, and 26,184 ‘Other’ phenotypes).
- When removing the locally-restricted samples, these now consist of 65,303 samples (28,586 PD cases, 15,258 Controls, and 21,459 ‘Other’ phenotypes).
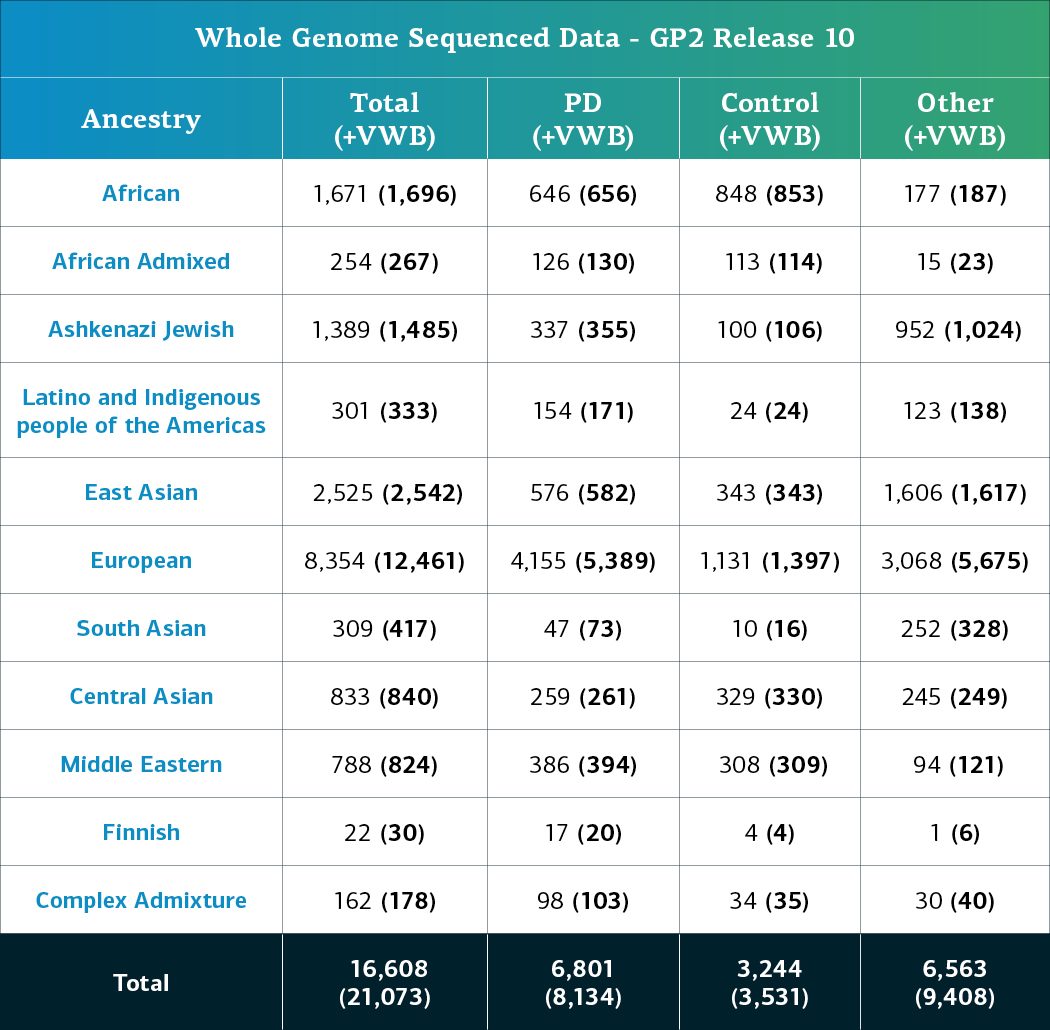
- The whole genome sequencing (WGS) data now consists of a total of 21,073 sequenced participants (8,134 PD cases, 3,531 Controls, and 9,408 ‘Other’ phenotypes).
- When removing the locally-restricted samples, these now consist of 16,608 participants (6,801 PD cases, 3,244 Controls, and 6,563 ‘Other’ phenotypes).
- Of note, cases recruited via the Monogenic network are coded as ‘Other’.
- The clinical exome data now consists of 10,454 samples with PD (Release 8).
- Of the 92,021 unique samples with genetic data (NBA, WGS, or clinical exome), 26,982 individuals also have additional extended clinical information.
What’s New In This Release?
Expanding Genomic Data
This release introduces a substantial expansion in the number of participants with available genetic data. We have added:
- 11,109 new participants with genotype array (NBA) data
- 13,339 new participants with whole genome sequencing (WGS) data
- 12,311 new participants with extended clinical data
- A family file (and corresponding data dictionary) which reports pairwise kinship estimates between individuals within families. It includes both inferred relationships (with kinship coefficients) and reported relationships.
Inclusion of PAR Region in Imputation
We’ve reintroduced the pseudoautosomal (PAR) region in the imputation of genotype array data, improving coverage and interpretation of sex chromosome variation. This enhancement is part of ongoing efforts to enhance genomic coverage and analytic accuracy.
Joint-calling Now Include AMP® PD cohorts
- The jointly-called WGS variant sets now include samples from the following five AMP® PD cohorts: BioFind, PPMI, LCC, STEADY-PD3 and SURE-PD3.
- By processing these samples together with GP2 rather than independently, it minimizes missingness, artifacts, and improves genotype accuracy.
- We have added a column to master key denoting which GP2 samples are also present in the AMP-PD dataset.
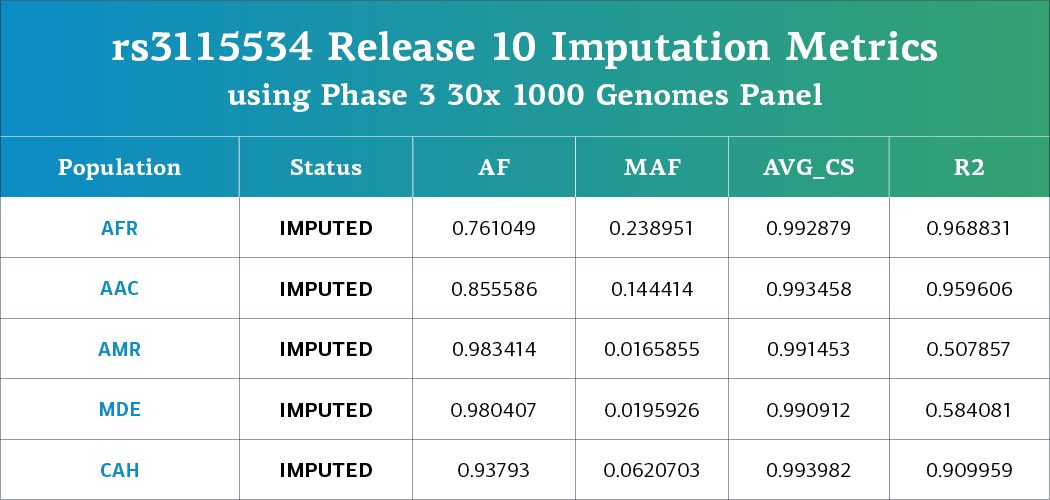
Targeted Imputation of rs3115534 Across Select Ancestries
In response to strong community interest in the intronic variant rs3115534, given that it’s been associated with increased risk of Parkinson’s disease and REM sleep behavior disorder, and has been functionally validated, we have now implemented a targeted imputation strategy to ensure its inclusion in the released datasets
- Specifically, chromosome 1 was imputed for five ancestries (AFR, AAC, AMR, MDE, and CAH) using the 1000 Genomes Phase 3 30x high coverage reference panel.
- Following imputation, data for rs3115534 was merged back into the TOPMed-based imputed files provided with GP2 releases. Note that imputation metrics for this variant did not meet quality thresholds (R² < 0.3) in other ancestry groups.
New Summary Statistics Now Available
We’ve made available several new GWAS summary statistics datasets, expanding global representation:
- GP2’s European (EUR) meta-GWAS (pre-print; GitHub)
- South African GWAS (pre-print pending; GitHub)
- Indian GWAS (pre-print; GitHub)
- RBD (REM Sleep Behavior Disorder) GWAS (pre-print pending; GitHub pending)
- LARGE-PD GWAS, which includes Latino American participants (pre-print pending; GitHub pending)
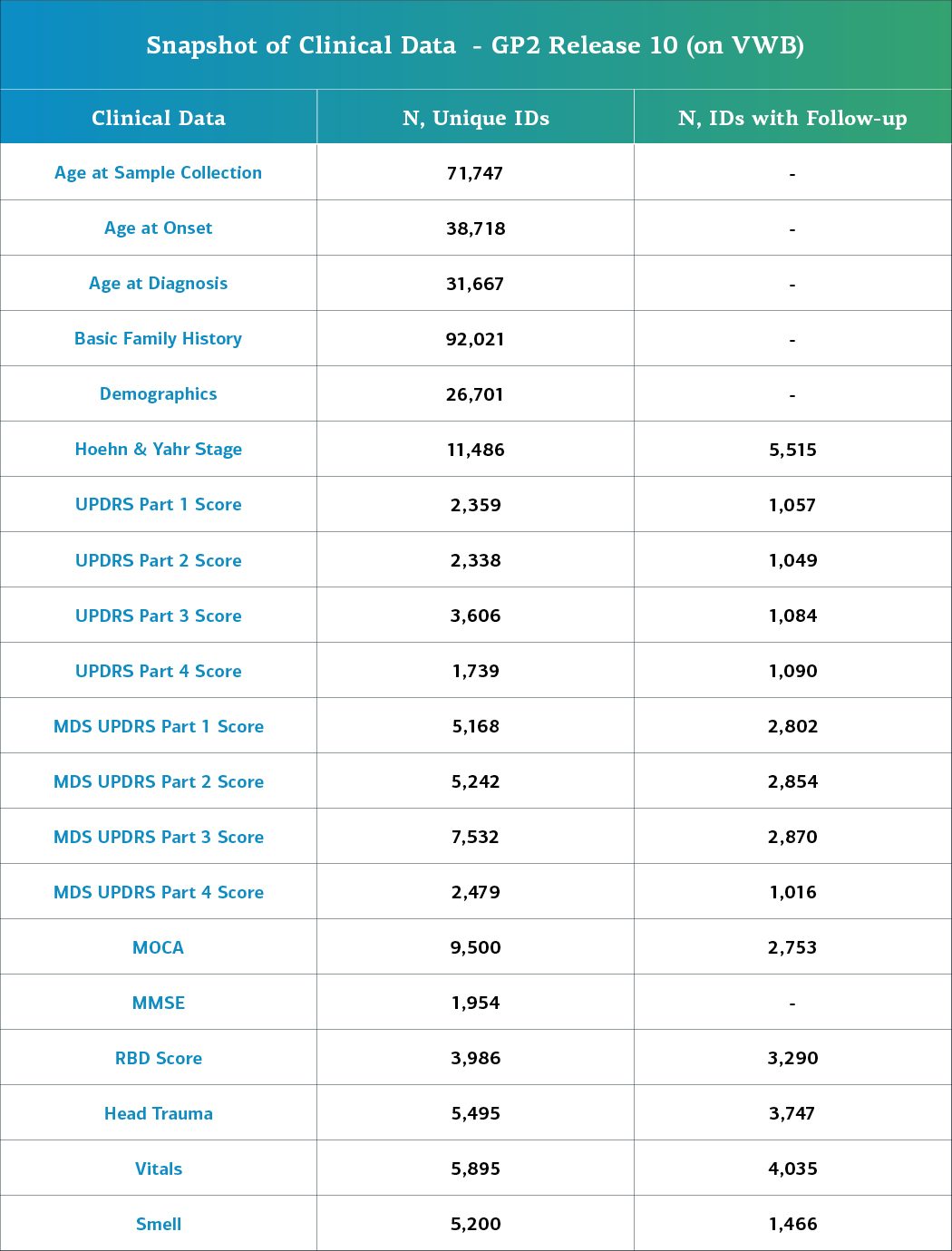
Clinical Data
This release contains clinical data for a total of 92,021 individuals who have genetic and core clinical data available. Of these, 26,982 have deep clinical phenotyping data available. This information consists of:
- Age at diagnosis and onset
- Primary, current, and latest diagnoses
- Cognitive exams such as the Mini-Mental State Examination (MMSE) and the Montreal Cognitive Assessment (MoCA)
- Movement Disorder Society-Sponsored Revision of the Unified Parkinson’s Disease Rating Scale (MDS-UPDRS)
- Detailed “other” phenotypes, such as Lewy body Dementia (LBD)
Individual-Level Data
We now capture the data from a total of 124 cohorts. Please refer to the GP2 Cohort Dashboard for more information on the cohorts that have been shared.
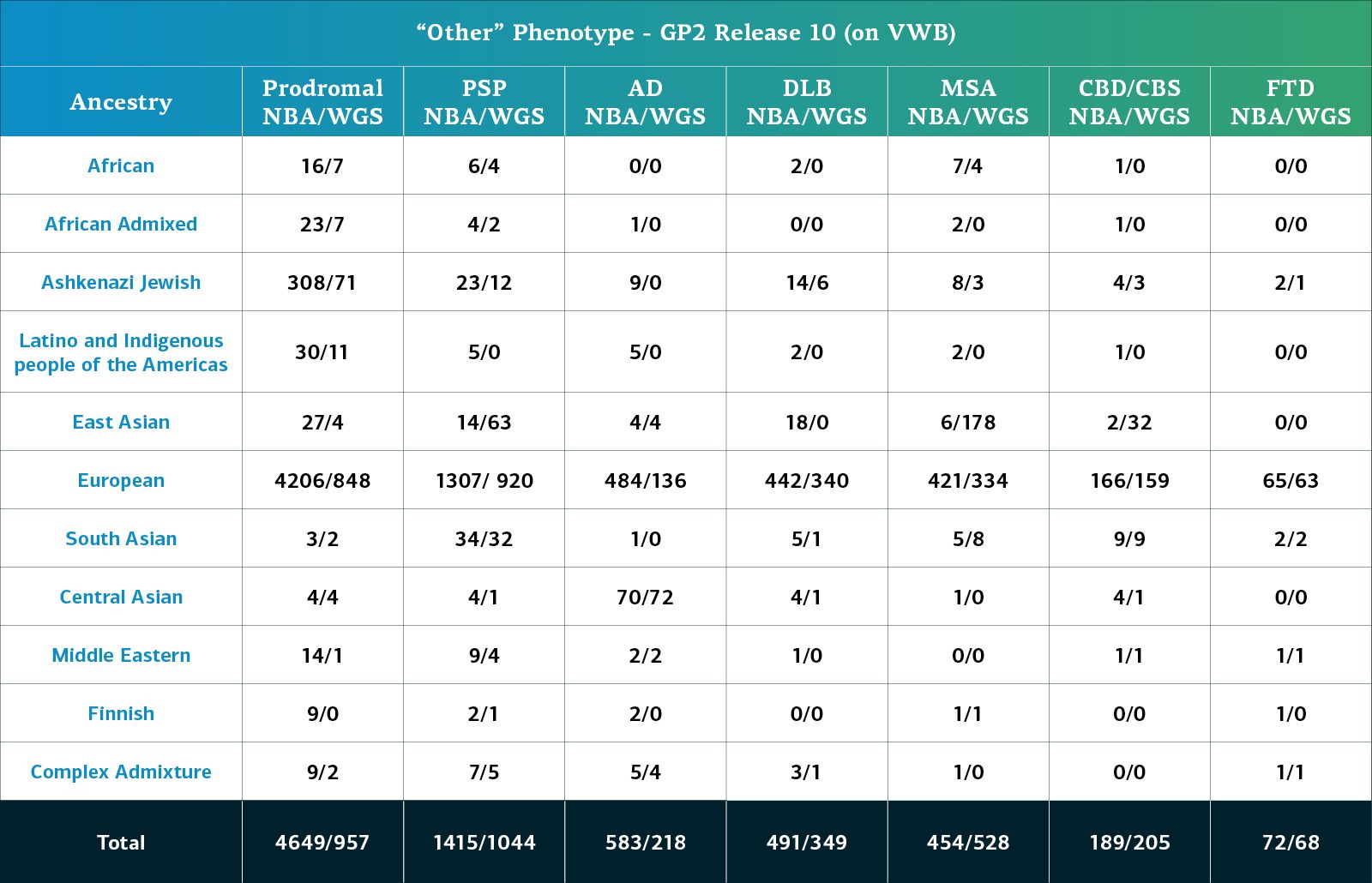
Genetically-determined ancestry of array genotyped GP2 participants are broken into 11 ancestry groups; the tables below provide details of the genetically-determined ancestry of participants in this release that have passed quality control for array data and whole genome sequencing data.. These numbers reflect samples from previous releases, reclustered using the updated cluster file and subjected to quality control, as well as newly genotyped samples exclusive to this release. The final table provides information about the genetically-determined ancestry of selected other, non-PD phenotypes.
Data Access
Locality-restricted GDPR samples via the Verily Viewpoint Workbench
We are continuing to pilot granting access to locally-restricted samples, otherwise known as samples governed by the General Data Protection Regulation (GDPR) policy, through our collaboration with the Verily Viewpoint Workbench.
At this time, as GP2 continues to roll out data sharing solutions for GDPR protected data, release 10 data with regional restrictions will be available to only GP2 consortium members and partners. As testing and implementation continues in 2025, this solution will be available to the broader research community. All release 10 samples can be found on Workbench, meanwhile all release 10 samples not governed by GDPR requirements can be found on the community workbench on Terra (like all previous releases). To gain access to the full release on VWB you must:
- Have approved GP2 Tier 2 access
- Fill out the GDPR-governed sample request form
- Be a GP2 consortium member (contributing cohort, GP2 partner, or project analyses team member)
Future data releases will continue to grow the diversity of participants available. You can check out our dashboard to see our progress. For users with tier 2 access already, you can explore the data further on our cohort browser, expanded on in a previous blog post.
As always, please refer to the README that accompanies each GP2 release for further details regarding recommendations for quality control, pipelines, data, and analyses!