We are proud to announce that the preprint of the next generation of Parkinson’s disease meta-analyses of genome-wide association studies (Meta-GWAS) by GP2 is now available.
The study is groundbreaking, encompassing 63,555 cases,17,700 proxy cases with a family history of Parkinson’s disease, and 1,746,386 controls – making it the most comprehensive investigation of Parkinson’s disease genetic risk to date. It highlights the power of open science, open data, and unified data generation and analysis processes deployed globally in the cloud.
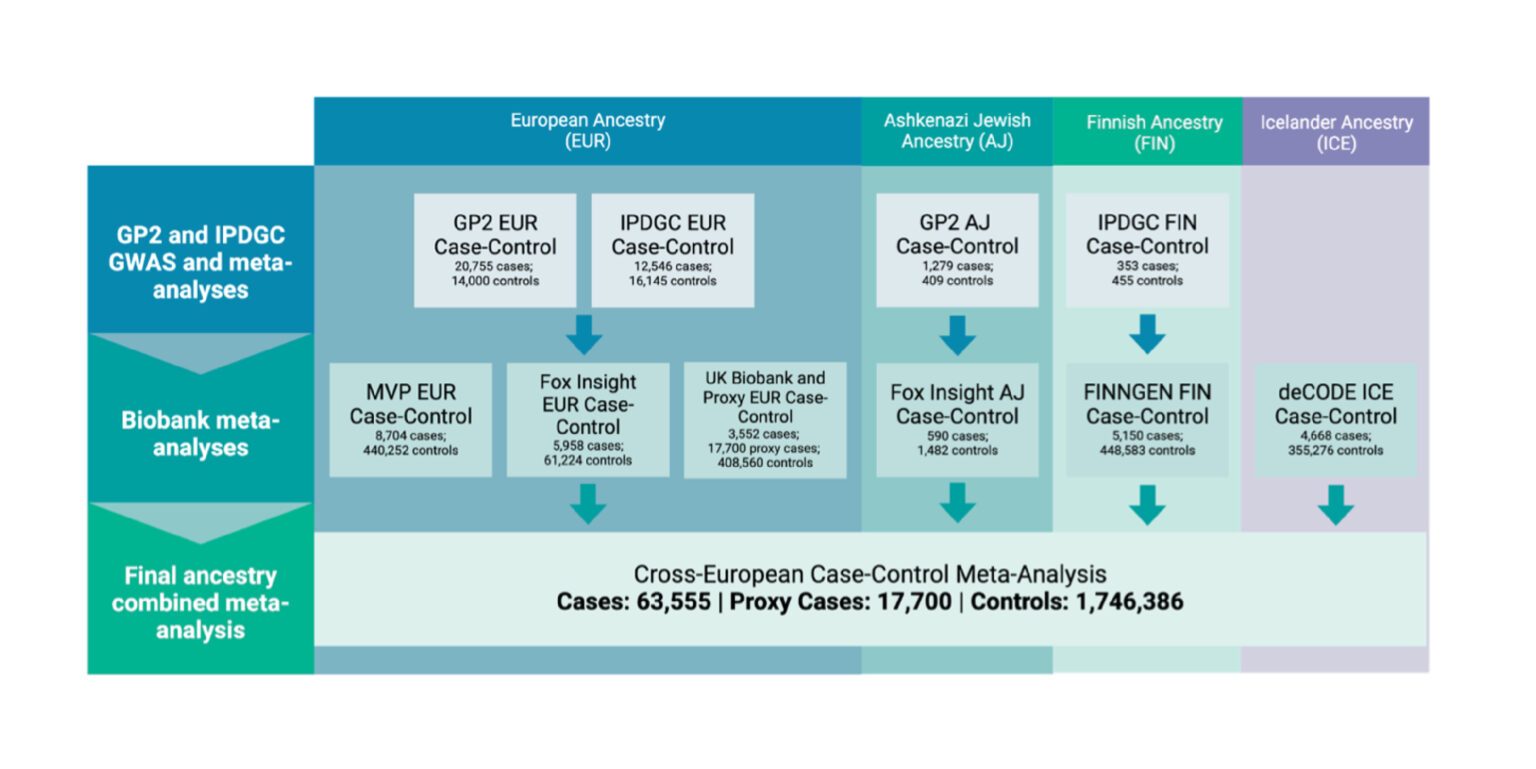
Key Study Findings
Our findings expand our understanding of Parkinson’s etiology, and include:
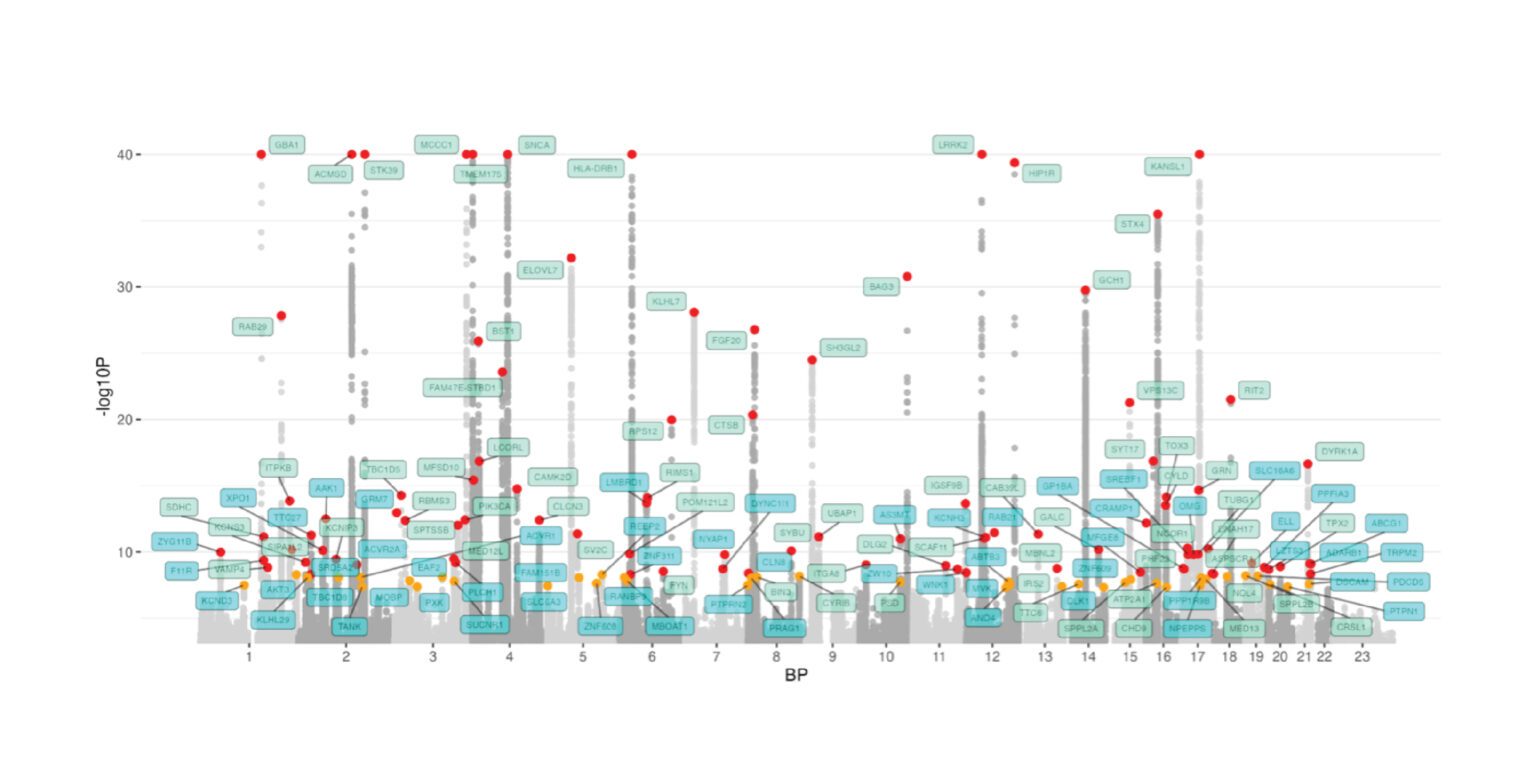
- Identification of 134 risk loci (59 novel) and a total of 157 independent risk variants across these loci, significantly expanding our understanding of Parkinson’s disease risk.
- Discovery through multi-omic data integration that the expression of the nominated risk genes is highly enriched in brain tissues, particularly in neuronal and astrocyte cell types.
- Prioritization of 33 high-confidence genes across the 134 loci for future follow-up studies.
Acknowledgments
This work is the result of a collaborative effort, with contributions from hundreds of individuals around the world. A few key individuals and their significant contributions include:
Hampton L. Leonard, Mary B. Makarious, Dan Vitale, and Mike A. Nalls designed the analysis, analyzed the data, and drafted the initial manuscript.
Astros Th. Skuladottir, Vala Palmadottir, Hreinn Stefansson, and Kari Stefansson designed the analysis and analyzed the data for the Iceland (ICE) ancestry GWAS from deCODE.
Kristin Levine, Hampton L. Leonard, Nicole Kuznetsov, Mary B. Makarious, Dan Vitale, Mat Koretsky contributed to quality control of GP2 genetic data and making it available to researchers.
Cornelis Blauwendraat and Andrew B. Singleton led study design, data logistics, and funding of the study.
Huw Morris, Manuela Tan, Hirotaka Iwaki, Simona Jasaityte, Ellie Stafford, Lietsel Jones, Shannon Ballard, J Solle, and Claire Wegel (Complex and Compliance Working Groups) designed the GP2 complex study, clinical and genetic data generation and preparation, and legal/data sharing logistics.
All other members of GP2 (contributors) contributed data and made critical revisions to this article.
A special thanks to Noel Burtt and her team from the Neurodegenerative Disease Knowledge Portal for hosting the summary statistics months in advance.