Explore GP2's Data Resources
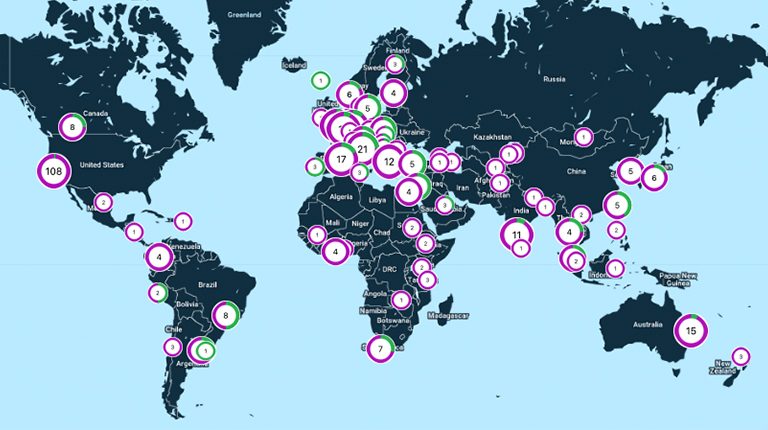

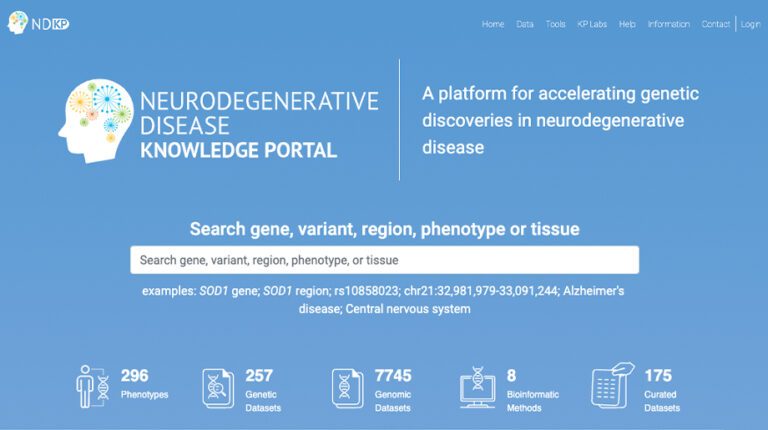
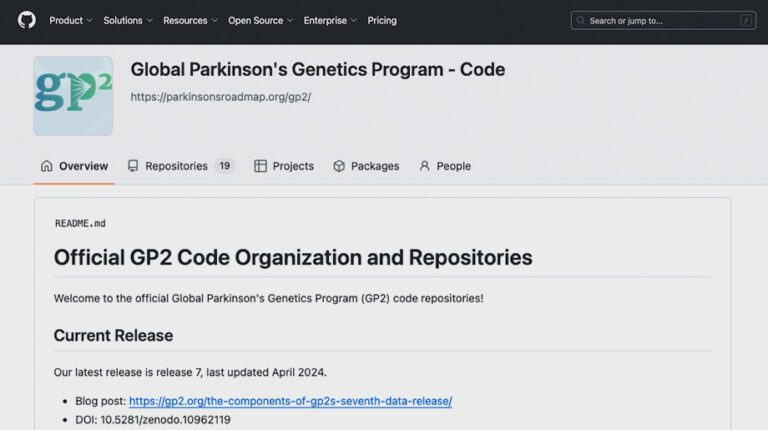




The Global Parkinson’s Genetics Program (GP2) is deeply committed to making science open, equipping a global research collective with the skill sets, tools, and infrastructure to rapidly and efficiently address emerging research needs and questions. Below is a snapshot summary of the GP2 resources that the community can leverage for their own research discovery.








