Overview
In December 2025, GP2 announced the 11th data release on the Terra and the Verily® Workbench platforms in collaboration with AMP® PD. This release includes 20,842 additional genotyped participants, 17,153 additional WGS participants, and 4,232 additional clinical exomes.
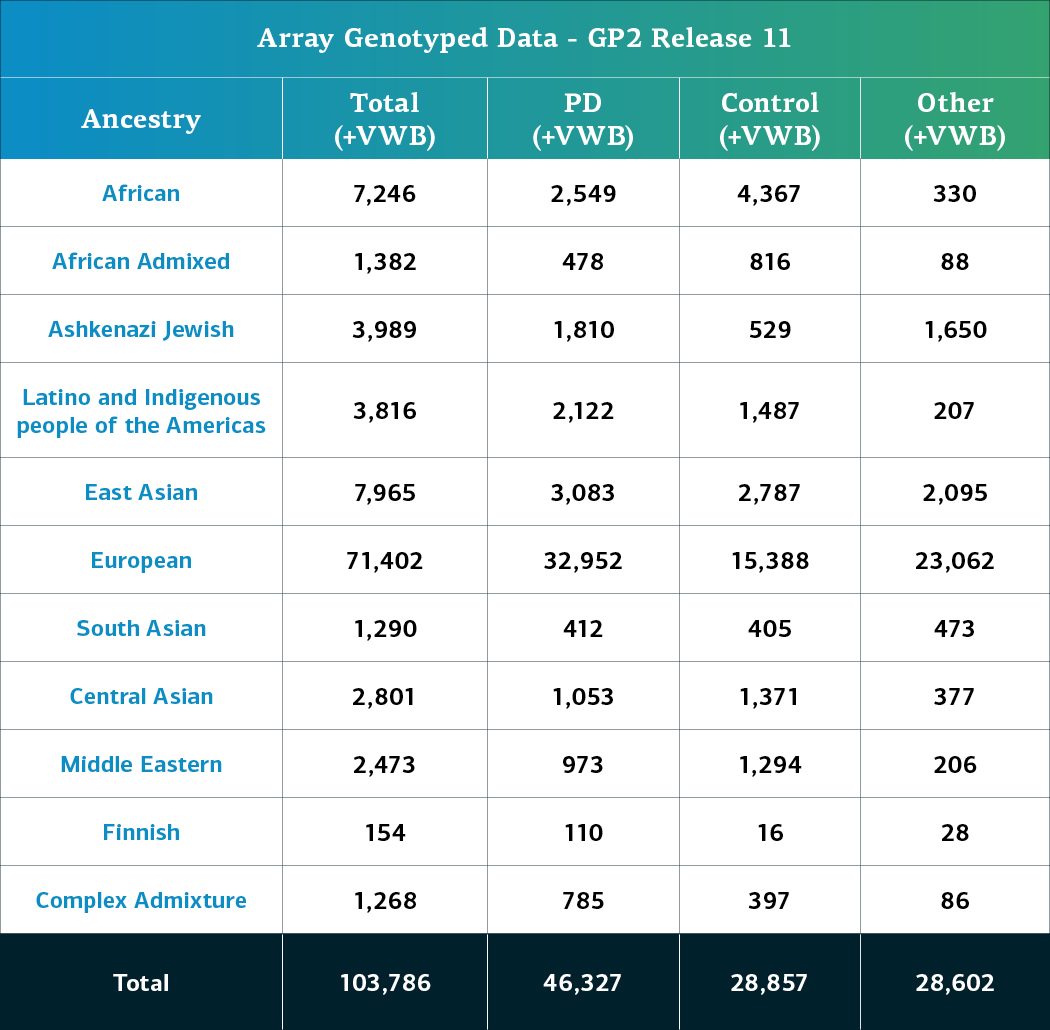
- The genotype array (NBA) data, including locally-restricted samples, now consists of a total of 103,786 genotyped participants (46,327 PD cases, 28,857 Controls, and 28,602 ‘Other’ phenotypes).
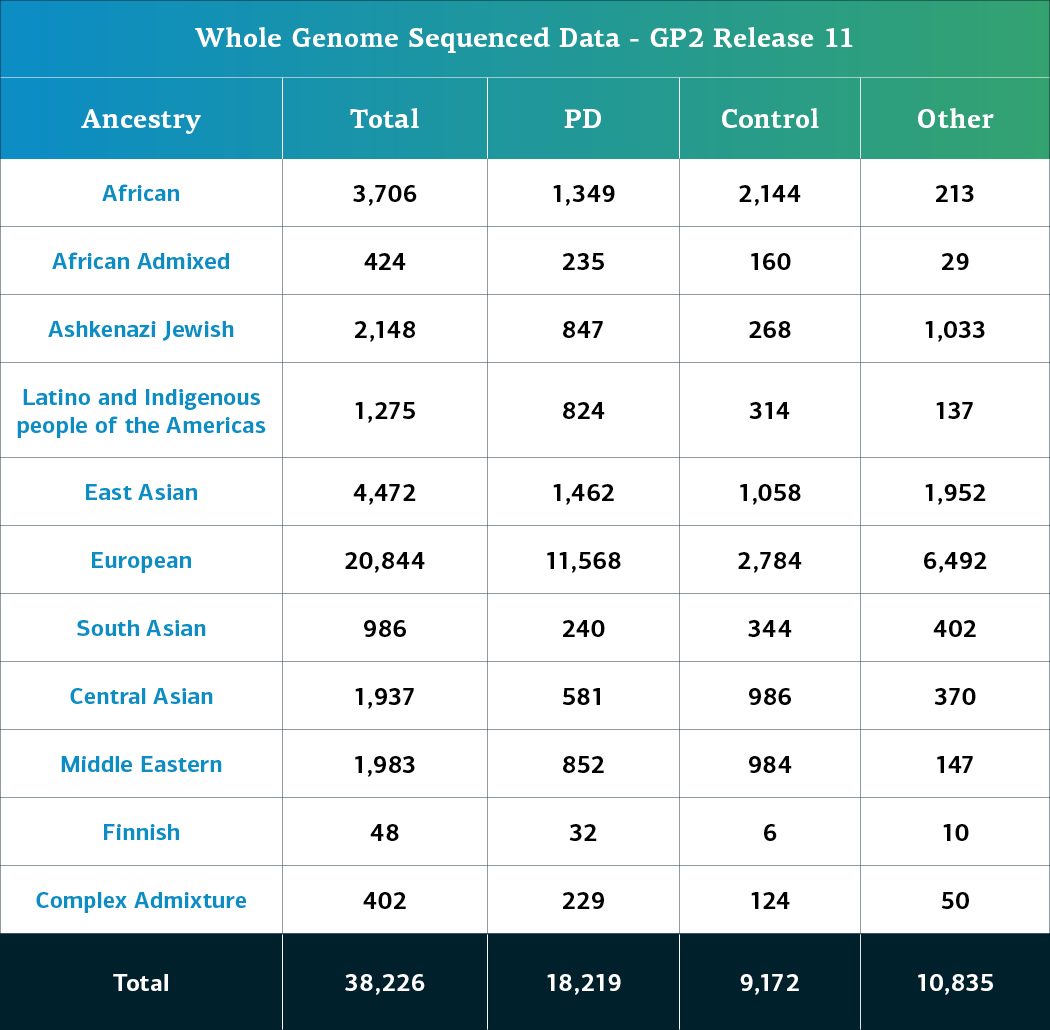
- The whole genome sequencing (WGS) data now consists of a total of 38,226 sequenced participants (18,219 PD cases, 9,172 Controls, and 10,835 ‘Other’ phenotypes).
- The clinical exome data now consists of 14,686 samples with PD.
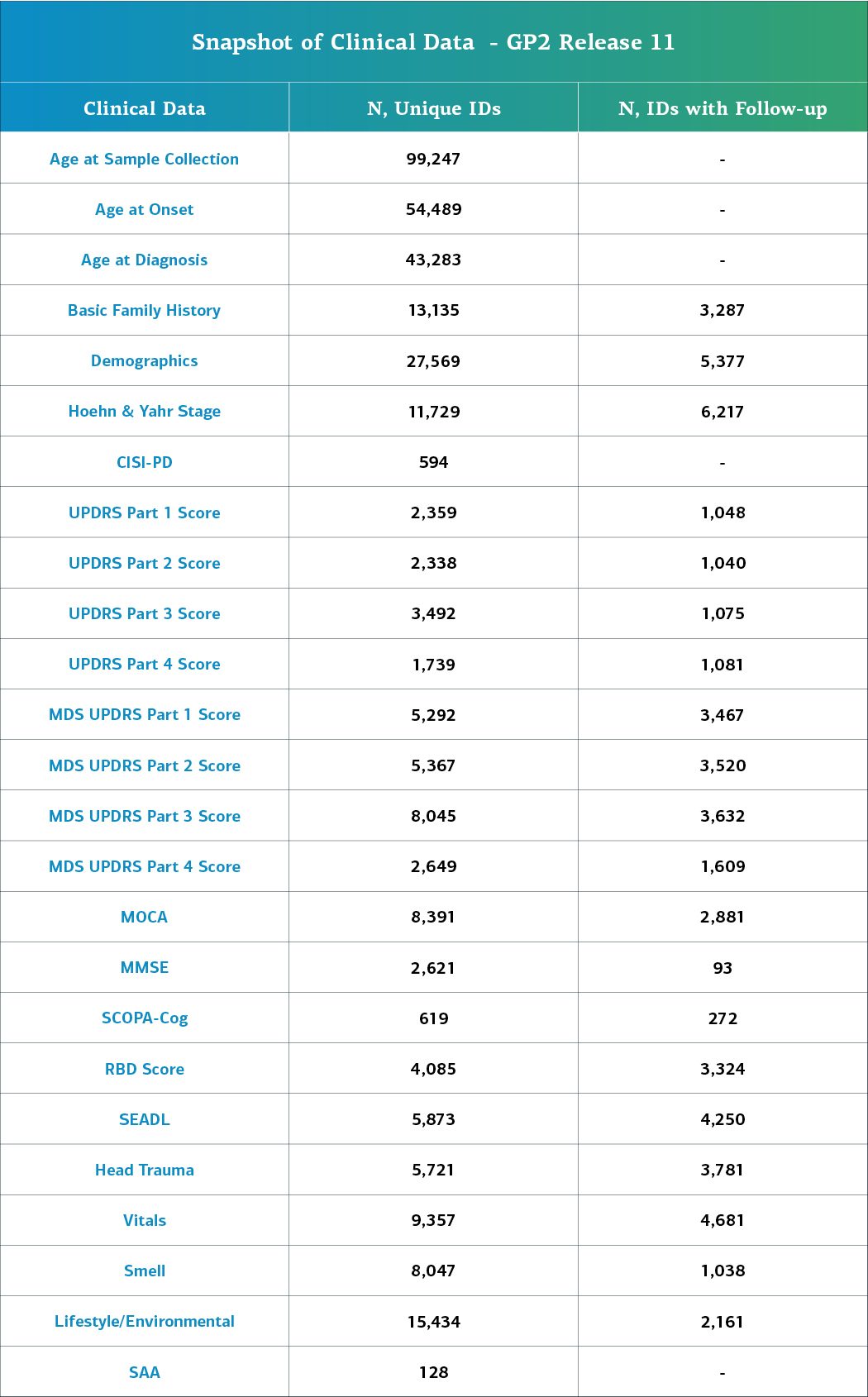
- Of the 122,317 unique samples with genetic data (NBA, WGS, or clinical exome), 32,897 individuals also have additional extended clinical information.
What’s New In This Release?
Expanding Genomic Data
This release introduces a substantial expansion in the number of participants with available genetic data. We have added:
- 20,842 new participants with genotype array (NBA) data
- 17,153 new participants with whole genome sequencing (WGS) data
- 5915 new participants with extended clinical data
- A family file (and corresponding data dictionary) which reports pairwise kinship estimates between individuals within families. It includes both inferred relationships (with kinship coefficients) and reported relationships.
Joint-calling Now Include AMP® PD cohorts
- The jointly-called WGS variant sets now include samples from the following seven AMP® PD cohorts: BioFIND, HBS, PDBP, PPMI, LCC, STEADY-PD3 and SURE-PD3.
- By processing these samples together with GP2 rather than independently, it minimizes missingness, artifacts, and improves genotype accuracy.
- We have added a column to master key denoting which GP2 samples are also present in the AMP-PD dataset.
New Summary Statistics Now Available
We’ve made available several additional GWAS summary statistics datasets under Tier 1:
- Blauwendraat et al PD age at onset (AAO) meta-analysis paper
- Blauwendraat et al PD autosomal sex-stratified meta-analysis paper
- Blauwendraat et al PD GBA1-modifier meta-analysis paper
Clinical Data
This release contains clinical data for a total of 122,317 individuals who have genetic and core clinical data available. Of these, 32,897 have deep clinical phenotyping data available. This information consists of:
- Age at diagnosis and onset
- Primary, current, and latest diagnoses
- Cognitive exams such as the Mini-Mental State Examination (MMSE) and the Montreal Cognitive Assessment (MoCA)
- Movement Disorder Society-Sponsored Revision of the Unified Parkinson’s Disease Rating Scale (MDS-UPDRS)
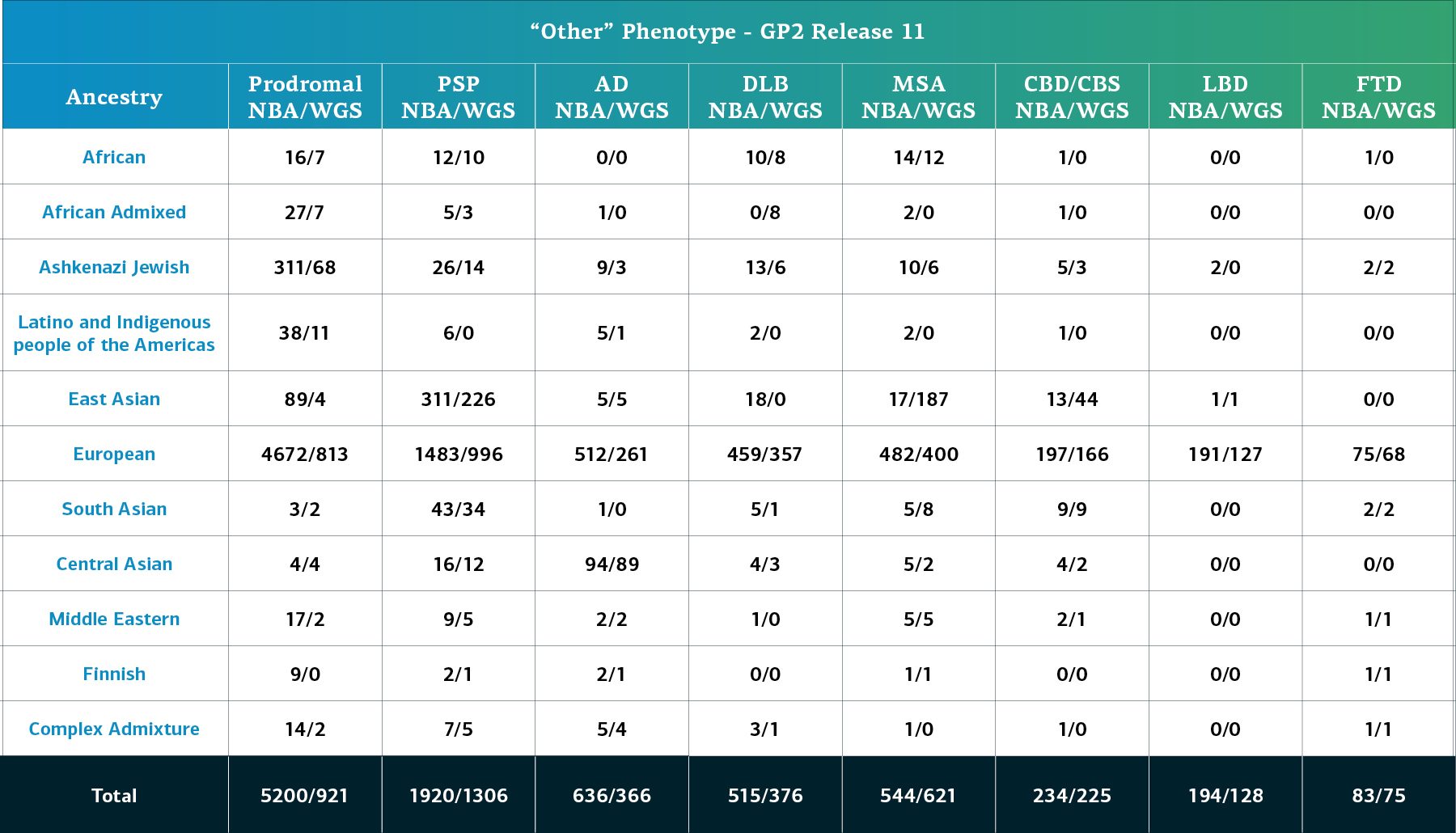
- Detailed “other” phenotypes, such as Lewy body Dementia (LBD)
Individual-Level Data
We now capture the data from a total of 148 cohorts. Please refer to the GP2 Cohort Dashboard for more information on the cohorts that have been shared.
Genetically-determined ancestry of array genotyped GP2 participants are broken into 11 ancestry groups; the tables below provide details of the genetically-determined ancestry of participants in this release that have passed quality control for array data and whole genome sequencing data. These numbers reflect samples from previous releases, reclustered using the updated cluster file and subjected to quality control, as well as newly genotyped samples exclusive to this release. The final table provides information about the genetically-determined ancestry of selected other, non-PD phenotypes.
Data Access
Locality-restricted GDPR samples via the Verily Viewpoint Workbench
We are continuing to pilot granting access to locally-restricted samples, otherwise known as samples governed by the General Data Protection Regulation (GDPR) policy, through our collaboration with the Verily Viewpoint Workbench. To gain access to the full release on VWB you must:
- Have approved GP2 Tier 2 access
- Fill out the GDPR-governed sample request form
Future data releases will continue to grow the diversity of participants available. You can check out our dashboard to see our progress. For users with tier 2 access already, you can explore the data further on our cohort browser, expanded on in a previous blog post.
As always, please refer to the README that accompanies each GP2 release for further details regarding recommendations for quality control, pipelines, data, and analyses!